DrC_00370
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00370 (1545 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P53634|CATC_HUMAN Dipeptidyl-peptidase I precursor (DPP-... 466 e-131 sp|P97821|CATC_MOUSE Dipeptidyl-peptidase I precursor (DPP-... 461 e-129 sp|Q60HG6|CATC_MACFA Dipeptidyl-peptidase I precursor (DPP-... 454 e-127 sp|P80067|CATC_RAT Dipeptidyl-peptidase I precursor (DPP-I)... 451 e-126 sp|O97578|CATC_CANFA Dipeptidyl-peptidase I precursor (DPP-... 444 e-124 sp|Q26563|CATC_SCHMA Cathepsin C precursor 425 e-118 sp|P80884|ANAN_ANACO Ananain precursor 152 3e-36 sp|P00786|CATH_RAT Cathepsin H precursor (Cathepsin B3) (Ca... 149 3e-35 sp|P25807|CPR1_CAEEL Gut-specific cysteine proteinase precu... 145 4e-34 sp|P49935|CATH_MOUSE Cathepsin H precursor (Cathepsin B3) (... 142 2e-33
>sp|P53634|CATC_HUMAN Dipeptidyl-peptidase I precursor (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase I exclusion domain chain; Dipeptidyl-peptidase I heavy chain; Dipeptidyl-peptidase I light chain] Length = 463 Score = 466 bits (1200), Expect = e-131 Identities = 235/469 (50%), Positives = 303/469 (64%), Gaps = 20/469 (4%) Frame = +3 Query: 96 AILLAFCLVQLSVS-----DTPANCTYQDVIGKWQVFTGNF----NVSCSTSKLVATKTL 248 ++LLA L+ LS DTPANCTY D++G W G+ +V+CS K + Sbjct: 6 SLLLAALLLLLSGDGAVRCDTPANCTYLDLLGTWVFQVGSSGSQRDVNCSVMGPQEKKVV 65 Query: 249 TLIYP-NWAVDEFGNYGKWTLIYNQGFEVTITNKKYFGFFDYKKINSTYTISYCDRLQPS 425 + + A D+ GN G +T+IYNQGFE+ + + K+F FF YK+ S T +YC+ Sbjct: 66 VYLQKLDTAYDDLGNSGHFTIIYNQGFEIVLNDYKWFAFFKYKEEGSKVT-TYCNETMTG 124 Query: 426 WFHDVLIRQWQCFKAQRTTTLKE---------KNNVLPHSNIFALTSLRYGSQKRIVDKI 578 W HDVL R W CF ++ T E KN+ +SN Y V I Sbjct: 125 WVHDVLGRNWACFTGKKVGTASENVYVNTAHLKNSQEKYSNRL------YKYDHNFVKAI 178 Query: 579 NLENNGWTAKDYPEFHEKTLYEVINMAGGSRSKLERPKPAPITKSILDSVKLIPKSFDWR 758 N WTA Y E+ TL ++I +GG K+ RPKPAP+T I + +P S+DWR Sbjct: 179 NAIQKSWTATTYMEYETLTLGDMIRRSGGHSRKIPRPKPAPLTAEIQQKILHLPTSWDWR 238 Query: 759 NVNGLNYVSPVRNQGGCGSCYSFASAGMLEARYRIRSNNTVRPILSPQDVVECSPYSQGC 938 NV+G+N+VSPVRNQ CGSCYSFAS GMLEAR RI +NN+ PILSPQ+VV CS Y+QGC Sbjct: 239 NVHGINFVSPVRNQASCGSCYSFASMGMLEARIRILTNNSQTPILSPQEVVSCSQYAQGC 298 Query: 939 DGGFPYLIAGKFAEDFGMAQESCNPYKGMNGKCSTTKDCKRYFATNYKYIGGYYGATNEP 1118 +GGFPYLIAGK+A+DFG+ +E+C PY G + C +DC RY+++ Y Y+GG+YG NE Sbjct: 299 EGGFPYLIAGKYAQDFGLVEEACFPYTGTDSPCKMKEDCFRYYSSEYHYVGGFYGGCNEA 358 Query: 1119 LMRMELVKNGPIAVGFEVYDDFMSYSGGVYHHNFGTRKLTTSKFGFNPFELTNHAVLVVG 1298 LM++ELV +GP+AV FEVYDDF+ Y G+YHH T + FNPFELTNHAVL+VG Sbjct: 359 LMKLELVHHGPMAVAFEVYDDFLHYKKGIYHH-------TGLRDPFNPFELTNHAVLLVG 411 Query: 1299 YG-ETSSGEKFWIVKNSWGNGWGENGYFRIRRGNDECGIESLGVASEPI 1442 YG +++SG +WIVKNSWG GWGENGYFRIRRG DEC IES+ VA+ PI Sbjct: 412 YGTDSASGMDYWIVKNSWGTGWGENGYFRIRRGTDECAIESIAVAATPI 460
>sp|P97821|CATC_MOUSE Dipeptidyl-peptidase I precursor (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase I exclusion domain chain; Dipeptidyl-peptidase I heavy chain; Dipeptidyl-peptidase I light chain] Length = 462 Score = 461 bits (1187), Expect = e-129 Identities = 232/465 (49%), Positives = 305/465 (65%), Gaps = 13/465 (2%) Frame = +3 Query: 87 MMLAILLAFCLVQLSVSDTPANCTYQDVIGKWQVFTG----NFNVSCSTSKLVATKTLTL 254 ++L +LL C V+ SDTPANCTY D++G W G +++CS + K + Sbjct: 11 VLLLVLLGVCTVR---SDTPANCTYPDLLGTWVFQVGPRSSRSDINCSVMEATEEKVVVH 67 Query: 255 IYP-NWAVDEFGNYGKWTLIYNQGFEVTITNKKYFGFFDYKKINSTYTISYCDRLQPSWF 431 + + A DE GN G +TLIYNQGFE+ + + K+F FF Y+ T ISYC W Sbjct: 68 LKKLDTAYDELGNSGHFTLIYNQGFEIVLNDYKWFAFFKYEVRGHT-AISYCHETMTGWV 126 Query: 432 HDVLIRQWQCFKAQRTTTLKEKNNVLPHSNIFALTSLR-------YGSQKRIVDKINLEN 590 HDVL R W CF ++ + EK N+ N L L+ Y V IN Sbjct: 127 HDVLGRNWACFVGKKVESHIEKVNM----NAAHLGGLQERYSERLYTHNHNFVKAINTVQ 182 Query: 591 NGWTAKDYPEFHEKTLYEVINMAGGSRSKLERPKPAPITKSILDSVKLIPKSFDWRNVNG 770 WTA Y E+ + +L ++I +G S+ ++ RPKPAP+T I + +P+S+DWRNV G Sbjct: 183 KSWTATAYKEYEKMSLRDLIRRSGHSQ-RIPRPKPAPMTDEIQQQILNLPESWDWRNVQG 241 Query: 771 LNYVSPVRNQGGCGSCYSFASAGMLEARYRIRSNNTVRPILSPQDVVECSPYSQGCDGGF 950 +NYVSPVRNQ CGSCYSFAS GMLEAR RI +NN+ PILSPQ+VV CSPY+QGCDGGF Sbjct: 242 VNYVSPVRNQESCGSCYSFASMGMLEARIRILTNNSQTPILSPQEVVSCSPYAQGCDGGF 301 Query: 951 PYLIAGKFAEDFGMAQESCNPYKGMNGKCSTTKDCKRYFATNYKYIGGYYGATNEPLMRM 1130 PYLIAGK+A+DFG+ +ESC PY + C ++C RY++++Y Y+GG+YG NE LM++ Sbjct: 302 PYLIAGKYAQDFGVVEESCFPYTAKDSPCKpreNCLRYYSSDYYYVGGFYGGCNEALMKL 361 Query: 1131 ELVKNGPIAVGFEVYDDFMSYSGGVYHHNFGTRKLTTSKFGFNPFELTNHAVLVVGYG-E 1307 ELVK+GP+AV FEV+DDF+ Y G+YHH T FNPFELTNHAVL+VGYG + Sbjct: 362 ELVKHGPMAVAFEVHDDFLHYHSGIYHH-------TGLSDPFNPFELTNHAVLLVGYGRD 414 Query: 1308 TSSGEKFWIVKNSWGNGWGENGYFRIRRGNDECGIESLGVASEPI 1442 +G ++WI+KNSWG+ WGE+GYFRIRRG DEC IES+ VA+ PI Sbjct: 415 PVTGIEYWIIKNSWGSNWGESGYFRIRRGTDECAIESIAVAAIPI 459
>sp|Q60HG6|CATC_MACFA Dipeptidyl-peptidase I precursor (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase I exclusion domain chain; Dipeptidyl-peptidase I heavy chain; Dipeptidyl-peptidase I light chain] Length = 463 Score = 454 bits (1169), Expect = e-127 Identities = 231/469 (49%), Positives = 297/469 (63%), Gaps = 20/469 (4%) Frame = +3 Query: 96 AILLAFCLVQLSVS-----DTPANCTYQDVIGKWQVFTGNF----NVSCSTSKLVATKTL 248 A LLA L+ LS DTPANCTY D++G W G+ +V+CS K + Sbjct: 6 ASLLAALLLLLSGDRAVRCDTPANCTYLDLLGTWVFQVGSSGSLRDVNCSVMGPPEKKVV 65 Query: 249 TLIYP-NWAVDEFGNYGKWTLIYNQGFEVTITNKKYFGFFDYKKINSTYTISYCDRLQPS 425 + + A D+ GN G +T+IYNQGFE+ + + K+F FF YK+ TI YC+ Sbjct: 66 VHLQKLDTAYDDLGNSGHFTIIYNQGFEIVLNDYKWFAFFKYKEEGIKVTI-YCNETMTG 124 Query: 426 WFHDVLIRQWQCFKAQRTTTLKE---------KNNVLPHSNIFALTSLRYGSQKRIVDKI 578 W HDVL R W CF ++ T E KN+ +SN Y V I Sbjct: 125 WVHDVLGRNWACFTGKKVGTASENVYVNTAHLKNSQEKYSNRL------YKYDHNFVKAI 178 Query: 579 NLENNGWTAKDYPEFHEKTLYEVINMAGGSRSKLERPKPAPITKSILDSVKLIPKSFDWR 758 N WTA Y E+ TL ++I +GG K+ RPKP P+T I + +P S+DWR Sbjct: 179 NAIQKSWTATTYMEYETLTLGDMIKRSGGHSRKIPRPKPTPLTAEIQQKILHLPTSWDWR 238 Query: 759 NVNGLNYVSPVRNQGGCGSCYSFASAGMLEARYRIRSNNTVRPILSPQDVVECSPYSQGC 938 NV+G+N+VSPVRNQ CGSCYSFAS GMLEAR RI +NN+ PILS Q+VV CS Y+QGC Sbjct: 239 NVHGINFVSPVRNQASCGSCYSFASVGMLEARIRILTNNSQTPILSSQEVVSCSQYAQGC 298 Query: 939 DGGFPYLIAGKFAEDFGMAQESCNPYKGMNGKCSTTKDCKRYFATNYKYIGGYYGATNEP 1118 +GGFPYL AGK+A+DFG+ +E+C PY G + C +DC RY+++ Y Y+GG+YG NE Sbjct: 299 EGGFPYLTAGKYAQDFGLVEEACFPYTGTDSPCKMKEDCFRYYSSEYHYVGGFYGGCNEA 358 Query: 1119 LMRMELVKNGPIAVGFEVYDDFMSYSGGVYHHNFGTRKLTTSKFGFNPFELTNHAVLVVG 1298 LM++ELV +GP+AV FEVYDDF+ Y G+YHH T + FNPFELTNHAVL+VG Sbjct: 359 LMKLELVYHGPLAVAFEVYDDFLHYQNGIYHH-------TGLRDPFNPFELTNHAVLLVG 411 Query: 1299 YG-ETSSGEKFWIVKNSWGNGWGENGYFRIRRGNDECGIESLGVASEPI 1442 YG +++SG +WIVKNSWG WGE+GYFRIRRG DEC IES+ VA+ PI Sbjct: 412 YGTDSASGMDYWIVKNSWGTSWGEDGYFRIRRGTDECAIESIAVAATPI 460
>sp|P80067|CATC_RAT Dipeptidyl-peptidase I precursor (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase I exclusion domain chain; Dipeptidyl-peptidase I heavy chain; Dipeptidyl-peptidase I light chain] Length = 462 Score = 451 bits (1160), Expect = e-126 Identities = 227/464 (48%), Positives = 298/464 (64%), Gaps = 13/464 (2%) Frame = +3 Query: 90 MLAILLAFCLVQLSVSDTPANCTYQDVIGKWQVFTG----NFNVSCSTSKLVATKTLTLI 257 +L +LL C V SDTPANCTY D++G W G +++CS + K + + Sbjct: 12 LLLVLLGVCTVS---SDTPANCTYPDLLGTWVFQVGPRHPRSHINCSVMEPTEEKVVIHL 68 Query: 258 YP-NWAVDEFGNYGKWTLIYNQGFEVTITNKKYFGFFDYKKINSTYTISYCDRLQPSWFH 434 + A DE GN G +TLIYNQGFE+ + + K+F FF Y+ + + ISYC W H Sbjct: 69 KKLDTAYDEVGNSGYFTLIYNQGFEIVLNDYKWFAFFKYE-VKGSRAISYCHETMTGWVH 127 Query: 435 DVLIRQWQCFKAQRTTTLKEKNNVLPHSNIFALTSLR-------YGSQKRIVDKINLENN 593 D L R W CF ++ EK V N+ L L+ Y V IN Sbjct: 128 DYLGRNWACFVGKKMANHSEKVYV----NVAHLGGLQEKYSERLYSHHHNFVKAINSVQK 183 Query: 594 GWTAKDYPEFHEKTLYEVINMAGGSRSKLERPKPAPITKSILDSVKLIPKSFDWRNVNGL 773 WTA Y + + ++ ++I +G S ++ RPKPAPIT I + +P+S+DWRNV G+ Sbjct: 184 SWTATTYRRYEKLSIRDLIRRSGHS-GRILRPKPAPITDEIQQQILSLPESWDWRNVRGI 242 Query: 774 NYVSPVRNQGGCGSCYSFASAGMLEARYRIRSNNTVRPILSPQDVVECSPYSQGCDGGFP 953 N+VSPVRNQ CGSCYSFAS GMLEAR RI +NN+ PILSPQ+VV CSPY+QGCDGGFP Sbjct: 243 NFVSPVRNQESCGSCYSFASIGMLEARIRILTNNSQTPILSPQEVVSCSPYAQGCDGGFP 302 Query: 954 YLIAGKFAEDFGMAQESCNPYKGMNGKCSTTKDCKRYFATNYKYIGGYYGATNEPLMRME 1133 YLIAGK+A+DFG+ +E+C PY + C ++C RY+++ Y Y+GG+YG NE LM++E Sbjct: 303 YLIAGKYAQDFGVVEENCFPYTATDAPCKPKENCLRYYSSEYYYVGGFYGGCNEALMKLE 362 Query: 1134 LVKNGPIAVGFEVYDDFMSYSGGVYHHNFGTRKLTTSKFGFNPFELTNHAVLVVGYG-ET 1310 LVK+GP+AV FEV+DDF+ Y G+YHH T FNPFELTNHAVL+VGYG + Sbjct: 363 LVKHGPMAVAFEVHDDFLHYHSGIYHH-------TGLSDPFNPFELTNHAVLLVGYGKDP 415 Query: 1311 SSGEKFWIVKNSWGNGWGENGYFRIRRGNDECGIESLGVASEPI 1442 +G +WIVKNSWG+ WGE+GYFRIRRG DEC IES+ +A+ PI Sbjct: 416 VTGLDYWIVKNSWGSQWGESGYFRIRRGTDECAIESIAMAAIPI 459
>sp|O97578|CATC_CANFA Dipeptidyl-peptidase I precursor (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase I exclusion domain chain; Dipeptidyl-peptidase I heavy chain 1; Dipeptidyl-peptidase I heavy chain 2; Dipeptidyl-peptidase I heavy chain 3; Dipeptidyl-peptidase I heavy chain 4; Dipeptidyl-peptidase I light chain] Length = 435 Score = 444 bits (1142), Expect = e-124 Identities = 225/448 (50%), Positives = 290/448 (64%), Gaps = 13/448 (2%) Frame = +3 Query: 138 DTPANCTYQDVIGKWQVF----TGNFNVSCSTSKLVATKTLTLIYP-NWAVDEFGNYGKW 302 DTPANCT+ +++G W VF G+ +V+CS K + + + A D FGN G + Sbjct: 1 DTPANCTHPELLGTW-VFQVGPAGSRSVNCSVMGPPEKKVVVHLEKLDTAYDNFGNTGHF 59 Query: 303 TLIYNQGFEVTITNKKYFGFFDYKKINSTYTISYCDRLQPSWFHDVLIRQWQCFKAQRTT 482 T+IYNQGFE+ + + K+F FF YK+ T SYC+ W HDVL R W CF + Sbjct: 60 TIIYNQGFEIVLNDYKWFAFFKYKEEGHKVT-SYCNETMTGWVHDVLGRNWACFTGTKMG 118 Query: 483 TLKEKNNV-------LPHSNIFALTSLRYGSQKRIVDKINLENNGWTAKDYPEFHEKTLY 641 T EK V L +N L Y V IN WTA Y E+ TL Sbjct: 119 TTSEKAKVNTKHIERLQENNSNRLYKYNY----EFVKAINTIQKSWTATRYIEYETLTLR 174 Query: 642 EVINMAGGSRSKLERPKPAPITKSILDSVKLIPKSFDWRNVNGLNYVSPVRNQGGCGSCY 821 +++ GG K+ RPKP P+T I + + +P S+DWRNV G N+VSPVRNQ CGSCY Sbjct: 175 DMMTRVGGR--KIPRPKPTPLTAEIHEEISRLPTSWDWRNVRGTNFVSPVRNQASCGSCY 232 Query: 822 SFASAGMLEARYRIRSNNTVRPILSPQDVVECSPYSQGCDGGFPYLIAGKFAEDFGMAQE 1001 +FAS MLEAR RI +NNT PILSPQ++V CS Y+QGC+GGFPYLIAGK+A+DFG+ +E Sbjct: 233 AFASTAMLEARIRILTNNTQTPILSPQEIVSCSQYAQGCEGGFPYLIAGKYAQDFGLVEE 292 Query: 1002 SCNPYKGMNGKCSTTKDCKRYFATNYKYIGGYYGATNEPLMRMELVKNGPIAVGFEVYDD 1181 +C PY G + C DC RY+++ Y Y+GG+YGA NE LM++ELV++GP+AV FEVYDD Sbjct: 293 ACFPYAGSDSPCK-PNDCFRYYSSEYYYVGGFYGACNEALMKLELVRHGPMAVAFEVYDD 351 Query: 1182 FMSYSGGVYHHNFGTRKLTTSKFGFNPFELTNHAVLVVGYG-ETSSGEKFWIVKNSWGNG 1358 F Y G+Y+H T + FNPFELTNHAVL+VGYG +++SG +WIVKNSWG+ Sbjct: 352 FFHYQKGIYYH-------TGLRDPFNPFELTNHAVLLVGYGTDSASGMDYWIVKNSWGSR 404 Query: 1359 WGENGYFRIRRGNDECGIESLGVASEPI 1442 WGE+GYFRIRRG DEC IES+ VA+ PI Sbjct: 405 WGEDGYFRIRRGTDECAIESIAVAATPI 432
>sp|Q26563|CATC_SCHMA Cathepsin C precursor Length = 454 Score = 425 bits (1093), Expect = e-118 Identities = 222/459 (48%), Positives = 291/459 (63%), Gaps = 10/459 (2%) Frame = +3 Query: 99 ILLAFCLVQLSVSDTPANCTYQDVIGKWQVFTGNFNVSCSTSKLVATKT--LTLIYPNWA 272 IL+ ++ + +DTPANCTY+D G+W+ G++ C KL + ++ ++L+YP+ A Sbjct: 8 ILIILACLRFTCADTPANCTYEDAHGRWKFHIGDYQSKCP-EKLNSKQSVVISLLYPDIA 66 Query: 273 VDEFGNYGKWTLIYNQGFEVTITNKKYFGFFDYKKINSTYTISYCDRLQPSWFHDVLIRQ 452 +DEFGN G WTLIYNQGFEVTI ++K+ F YK N + C + P W HD LI Sbjct: 67 IDEFGNRGHWTLIYNQGFEVTINHRKWLVIFAYKS-NGEFN---CHKSMPMWTHDTLIDS 122 Query: 453 WQCFKAQRTTTLKEKNNVLPHSNIFALTSLRYGSQKRIVDKINLENNGWTAKDYPEFHEK 632 + K N L S F T Y V KIN W + YPE + Sbjct: 123 GSVCSGKIGVHDKFHINKLFGSKSFGRTL--YHINPSFVGKINAHQKSWRGEIYPELSKY 180 Query: 633 TLYEVINMAGGSRSKLERP----KPAPITKSILDSVKLIPKSFDWRNV--NGLNYVSPVR 794 T+ E+ N AGG +S + RP + P +K ++ +P FDW + + V+P+R Sbjct: 181 TIDELRNRAGGVKSMVTRPSVLNRKTP-SKELISLTGNLPLEFDWTSPPDGSRSPVTPIR 239 Query: 795 NQGGCGSCYSFASAGMLEARYRIRSNNTVRPILSPQDVVECSPYSQGCDGGFPYLIAGKF 974 NQG CGSCY+ SA LEAR R+ SN + +PILSPQ VV+CSPYS+GC+GGFP+LIAGK+ Sbjct: 240 NQGICGSCYASPSAAALEARIRLVSNFSEQPILSPQTVVDCSPYSEGCNGGFPFLIAGKY 299 Query: 975 AEDFGMAQESCNPYKGMN-GKCSTTKDCKRYFATNYKYIGGYYGATNEPLMRMELVKNGP 1151 EDFG+ Q+ PY G + GKC+ +K+C RY+ T+Y YIGGYYGATNE LM++EL+ NGP Sbjct: 300 GEDFGLPQKIVIPYTGEDTGKCTVSKNCTRYYTTDYSYIGGYYGATNEKLMQLELISNGP 359 Query: 1152 IAVGFEVYDDFMSYSGGVYHHNFGTRKLTTSKFGFNPFELTNHAVLVVGYG-ETSSGEKF 1328 VGFEVY+DF Y G+YHH + T + FNPFELTNHAVL+VGYG + SGE + Sbjct: 360 FPVGFEVYEDFQFYKEGIYHHT----TVQTDHYNFNPFELTNHAVLLVGYGVDKLSGEPY 415 Query: 1329 WIVKNSWGNGWGENGYFRIRRGNDECGIESLGVASEPIL 1445 W VKNSWG WGE GYFRI RG DECG+ESLGV +P+L Sbjct: 416 WKVKNSWGVEWGEQGYFRILRGTDECGVESLGVRFDPVL 454
>sp|P80884|ANAN_ANACO Ananain precursor Length = 345 Score = 152 bits (383), Expect = 3e-36 Identities = 104/346 (30%), Positives = 161/346 (46%) Frame = +3 Query: 405 CDRLQPSWFHDVLIRQWQCFKAQRTTTLKEKNNVLPHSNIFALTSLRYGSQKRIVDKINL 584 CD +PS D +++Q++ + A+ K+ + + IF + ++ N Sbjct: 26 CD--EPS---DPMMKQFEEWMAEYGRVYKDNDEKMLRFQIFK-------NNVNHIETFNN 73 Query: 585 ENNGWTAKDYPEFHEKTLYEVINMAGGSRSKLERPKPAPITKSILDSVKLIPKSFDWRNV 764 N +F + T E + G L + ++ +D + +P+S DWR+ Sbjct: 74 RNGNSYTLGINQFTDMTNNEFVAQYTGLSLPLNIKREPVVSFDDVD-ISSVPQSIDWRDS 132 Query: 765 NGLNYVSPVRNQGGCGSCYSFASAGMLEARYRIRSNNTVRPILSPQDVVECSPYSQGCDG 944 V+ V+NQG CGSC++FAS +E+ Y+I+ N V LS Q V++C+ S GC G Sbjct: 133 GA---VTSVKNQGRCGSCWAFASIATVESIYKIKRGNLVS--LSEQQVLDCA-VSYGCKG 186 Query: 945 GFPYLIAGKFAEDFGMAQESCNPYKGMNGKCSTTKDCKRYFATNYKYIGGYYGATNEPLM 1124 G+ + G+A + PYK G C T + T Y Y+ N Sbjct: 187 GWINKAYSFIISNKGVASAAIYPYKAAKGTCKTNGVPNSAYITRYTYV-----QRNNERN 241 Query: 1125 RMELVKNGPIAVGFEVYDDFMSYSGGVYHHNFGTRKLTTSKFGFNPFELTNHAVLVVGYG 1304 M V N PIA + +F Y GV+ GTR NHA++++GYG Sbjct: 242 MMYAVSNQPIAAALDASGNFQHYKRGVFTGPCGTR--------------LNHAIVIIGYG 287 Query: 1305 ETSSGEKFWIVKNSWGNGWGENGYFRIRRGNDECGIESLGVASEPI 1442 + SSG+KFWIV+NSWG GWGE GY R+ R + G+A +P+ Sbjct: 288 QDSSGKKFWIVRNSWGAGWGEGGYIRLAR-DVSSSFGLCGIAMDPL 332
>sp|P00786|CATH_RAT Cathepsin H precursor (Cathepsin B3) (Cathepsin BA) [Contains: Cathepsin H mini chain; Cathepsin H heavy chain; Cathepsin H light chain] Length = 333 Score = 149 bits (375), Expect = 3e-35 Identities = 98/255 (38%), Positives = 127/255 (49%), Gaps = 3/255 (1%) Frame = +3 Query: 687 PKPAPITKS-ILDSVKLIPKSFDWRNVNGLNYVSPVRNQGGCGSCYSFASAGMLEARYRI 863 P+ TKS L P S DWR N VSPV+NQG CGSC++F++ G LE+ I Sbjct: 97 PQNCSATKSNYLRGTGPYPSSMDWRKKG--NVVSPVKNQGACGSCWTFSTTGALESAVAI 154 Query: 864 RSNNTVRPILSPQDVVECSPY--SQGCDGGFPYLIAGKFAEDFGMAQESCNPYKGMNGKC 1037 S + L+ Q +V+C+ + GC GG P + G+ E PY G NG+C Sbjct: 155 ASGKMMT--LAEQQLVDCAQNFNNHGCQGGLPSQAFEYILYNKGIMGEDSYPYIGKNGQC 212 Query: 1038 STTKDCKRYFATNYKYIGGYYGATNEPLMRMELVKNGPIAVGFEVYDDFMSYSGGVYHHN 1217 + F N I +E M + P++ FEV +DFM Y GVY N Sbjct: 213 KFNPEKAVAFVKNVVNIT----LNDEAAMVEAVALYNPVSFAFEVTEDFMMYKSGVYSSN 268 Query: 1218 FGTRKLTTSKFGFNPFELTNHAVLVVGYGETSSGEKFWIVKNSWGNGWGENGYFRIRRGN 1397 + T K NHAVL VGYGE +G +WIVKNSWG+ WG NGYF I RG Sbjct: 269 SCHK--TPDK--------VNHAVLAVGYGE-QNGLLYWIVKNSWGSNWGNNGYFLIERGK 317 Query: 1398 DECGIESLGVASEPI 1442 + CG+ + AS PI Sbjct: 318 NMCGLAA--CASYPI 330
>sp|P25807|CPR1_CAEEL Gut-specific cysteine proteinase precursor Length = 329 Score = 145 bits (365), Expect = 4e-34 Identities = 88/257 (34%), Positives = 134/257 (52%), Gaps = 23/257 (8%) Frame = +3 Query: 735 IPKSFDWRNV-NGLNYVSPVRNQGGCGSCYSFASAGMLEARYRIRSNNTVRPILSPQDVV 911 +P +FD R + + +R+Q CGSC++F +A M+ R I + +PI+SP D++ Sbjct: 85 VPATFDSRTQWSECKSIKLIRDQATCGSCWAFGAAEMISDRTCIETKGAQQPIISPDDLL 144 Query: 912 EC--SPYSQGCDGGFPYLIAGKFAEDFGMAQ------ESCNPYK---GMNGKCSTTK--- 1049 C S GC+GG+P + A ++ + G+ C PY +G C +K Sbjct: 145 SCCGSSCGNGCEGGYP-IQALRWWDSKGVVTGGDYHGAGCKPYPIAPCTSGNCPESKTPS 203 Query: 1050 ---DCKRYFATNY---KYIG--GYYGATNEPLMRMELVKNGPIAVGFEVYDDFMSYSGGV 1205 C+ ++T Y K+ G Y N ++ E+ NGP+ F VY+DF Y GV Sbjct: 204 CSMSCQSGYSTAYAKDKHFGVSAYAVPKNAASIQAEIYANGPVEAAFSVYEDFYKYKSGV 263 Query: 1206 YHHNFGTRKLTTSKFGFNPFELTNHAVLVVGYGETSSGEKFWIVKNSWGNGWGENGYFRI 1385 Y H T K+ L HA+ ++G+G T SG +W+V NSWG WGE+G+F+I Sbjct: 264 YKH-------TAGKY------LGGHAIKIIGWG-TESGSPYWLVANSWGVNWGESGFFKI 309 Query: 1386 RRGNDECGIESLGVASE 1436 RG+D+CGIES VA + Sbjct: 310 YRGDDQCGIESAVVAGK 326
>sp|P49935|CATH_MOUSE Cathepsin H precursor (Cathepsin B3) (Cathepsin BA) [Contains: Cathepsin H mini chain; Cathepsin H heavy chain; Cathepsin H light chain] Length = 333 Score = 142 bits (359), Expect = 2e-33 Identities = 94/255 (36%), Positives = 125/255 (49%), Gaps = 3/255 (1%) Frame = +3 Query: 687 PKPAPITKS-ILDSVKLIPKSFDWRNVNGLNYVSPVRNQGGCGSCYSFASAGMLEARYRI 863 P+ TKS L P S DWR N VSPV+NQG C SC++F++ G LE+ I Sbjct: 97 PQNCSATKSNYLRGTGPYPSSMDWRKKG--NVVSPVKNQGACASCWTFSTTGALESAVAI 154 Query: 864 RSNNTVRPILSPQDVVECSPY--SQGCDGGFPYLIAGKFAEDFGMAQESCNPYKGMNGKC 1037 S + L+ Q +V+C+ + GC GG P + G+ +E PY G + C Sbjct: 155 ASGKMLS--LAEQQLVDCAQAFNNHGCKGGLPSQAFEYILYNKGIMEEDSYPYIGKDSSC 212 Query: 1038 STTKDCKRYFATNYKYIGGYYGATNEPLMRMELVKNGPIAVGFEVYDDFMSYSGGVYHHN 1217 F N I +E M + P++ FEV +DF+ Y GVY Sbjct: 213 RFNPQKAVAFVKNVVNIT----LNDEAAMVEAVALYNPVSFAFEVTEDFLMYKSGVY--- 265 Query: 1218 FGTRKLTTSKFGFNPFELTNHAVLVVGYGETSSGEKFWIVKNSWGNGWGENGYFRIRRGN 1397 +SK + NHAVL VGYGE +G +WIVKNSWG+ WGENGYF I RG Sbjct: 266 -------SSKSCHKTPDKVNHAVLAVGYGE-QNGLLYWIVKNSWGSQWGENGYFLIERGK 317 Query: 1398 DECGIESLGVASEPI 1442 + CG+ + AS PI Sbjct: 318 NMCGLAA--CASYPI 330
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 183,410,274
Number of Sequences: 369166
Number of extensions: 4046476
Number of successful extensions: 10738
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9509
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10182
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 18797071545
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail
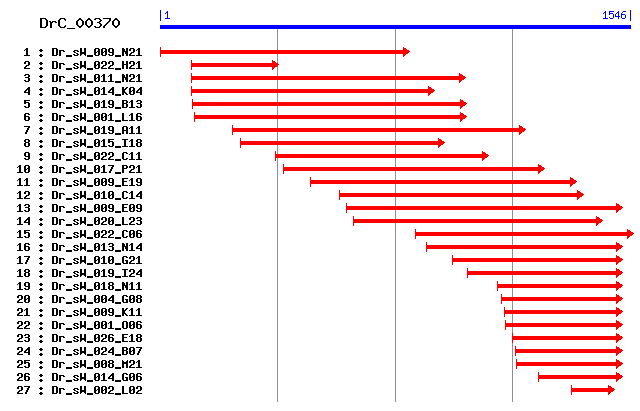
- Dr_sW_022_H21
- Dr_sW_009_N21
- Dr_sW_014_K04
- Dr_sW_015_I18
- Dr_sW_011_N21
- Dr_sW_019_B13
- Dr_sW_001_L16
- Dr_sW_022_C11
- Dr_sW_019_A11
- Dr_sW_017_P21
- Dr_sW_009_E19
- Dr_sW_010_C14
- Dr_sW_002_L02
- Dr_sW_020_L23
- Dr_sW_009_E09
- Dr_sW_014_G06
- Dr_sW_009_K11
- Dr_sW_024_B07
- Dr_sW_010_G21
- Dr_sW_001_O06
- Dr_sW_013_N14
- Dr_sW_018_N11
- Dr_sW_019_I24
- Dr_sW_022_C06
- Dr_sW_008_M21
- Dr_sW_004_G08
- Dr_sW_026_E18