DrC_00849
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00849 (2296 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P02828|HSP83_DROME Heat shock protein 83 (HSP 82) 983 0.0 sp|P11501|HS90A_CHICK Heat shock protein HSP 90-alpha 982 0.0 sp|P08238|HS90B_HUMAN Heat shock protein HSP 90-beta (HSP 8... 975 0.0 sp|Q9GKX8|HS90B_HORSE Heat shock protein HSP 90-beta (HSP 84) 974 0.0 sp|O57521|HS90B_BRARE Heat shock protein HSP 90-beta 971 0.0 sp|P11499|HS90B_MOUSE Heat shock protein HSP 90-beta (HSP 8... 970 0.0 sp|P34058|HS90B_RAT Heat shock protein HSP 90-beta (HSP 84) 957 0.0 sp|O61998|HSP90_BRUPA Heat shock protein 90 671 0.0 sp|P07900|HS90A_HUMAN Heat shock protein HSP 90-alpha (HSP 86) 669 0.0 sp|O02705|HS90A_PIG Heat shock protein HSP 90-alpha (HSP 86) 667 0.0
>sp|P02828|HSP83_DROME Heat shock protein 83 (HSP 82) Length = 717 Score = 983 bits (2541), Expect = 0.0 Identities = 499/715 (69%), Positives = 571/715 (79%), Gaps = 2/715 (0%) Frame = +2 Query: 44 DTETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYKSLTDPSVLDTAKE 223 + ETFAFQAEIAQLMSLIINTFYSNKEIFLRELISN+SDALDKIRY+SLTDPS LD+ KE Sbjct: 4 EAETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNASDALDKIRYESLTDPSKLDSGKE 63 Query: 224 LMIKVIPDKDAGTLTIIDSGNGMTKADLINNLGTIARSGTKAFMEALQAGADISMIGQFG 403 L IK+IP+K AGTLTIID+G GMTK+DL+NNLGTIA+SGTKAFMEALQAGADISMIGQFG Sbjct: 64 LYIKLIPNKTAGTLTIIDTGIGMTKSDLVNNLGTIAKSGTKAFMEALQAGADISMIGQFG 123 Query: 404 VGFYSSYLVADKVQVISKSNDDEQYLWESSAGGSFTIKNDNSEPLGRGTKIILFMKEDQV 583 VGFYS+YLVADKV V SK+NDDEQY+WESSAGGSFT++ DNSEPLGRGTKI+L++KEDQ Sbjct: 124 VGFYSAYLVADKVTVTSKNNDDEQYVWESSAGGSFTVRADNSEPLGRGTKIVLYIKEDQT 183 Query: 584 EYLEERKIKDIIKKHSQFIGYPIKLVLEKERXXXXXXXXXXXXXXXXXXXXXXXXXXXXX 763 +YLEE KIK+I+ KHSQFIGYPIKL++EKER Sbjct: 184 DYLEESKIKEIVNKHSQFIGYPIKLLVEKEREKEVSDDEADDEKKEGDEKKEMETDEPKI 243 Query: 764 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXLNKTKPIWTRNPDDVSQEDYAEFYKSLTN 943 LNKTKPIWTRNPDD+SQE+Y EFYKSLTN Sbjct: 244 EDVGEDEDADKKDKDAKKKKTIKEKYTEDEELNKTKPIWTRNPDDISQEEYGEFYKSLTN 303 Query: 944 DWEDHLAVKHFSVEGQLEFRALLFCPKRAPFDLFENKKKRNNIKLYVRRVFIMDNCEDLI 1123 DWEDHLAVKHFSVEGQLEFRALLF P+R PFDLFEN+KKRNNIKLYVRRVFIMDNCEDLI Sbjct: 304 DWEDHLAVKHFSVEGQLEFRALLFIPRRTPFDLFENQKKRNNIKLYVRRVFIMDNCEDLI 363 Query: 1124 PEYLNFVKGVVDSEDLPLNISREMLQQNKILKVIRKNLVKKCIELFEEIMEDKDNYKKFY 1303 PEYLNF+KGVVDSEDLPLNISREMLQQNK+LKVIRKNLVKK +EL EE+ EDK+NYKKFY Sbjct: 364 PEYLNFMKGVVDSEDLPLNISREMLQQNKVLKVIRKNLVKKTMELIEELTEDKENYKKFY 423 Query: 1304 EQFSKNIKLGIHEDSVNRNKIAEFLRYYSTSSGDEMCSLKDYVSRMKENQKDIYYITGES 1483 +QFSKN+KLG+HEDS NR K+A+FLR+++++SGD+ CSL DYVSRMK+NQK +Y+ITGES Sbjct: 424 DQFSKNLKLGVHEDSNNRAKLADFLRFHTSASGDDFCSLADYVSRMKDNQKHVYFITGES 483 Query: 1484 KDNVNHSAFVERLTKRGFEVLLMVDPIDEYSVQQLKEYDGKKLVCVTXXXXXXXXXXXXX 1663 KD V++SAFVER+ RGFEV+ M +PIDEY +Q LKEY GK+LV VT Sbjct: 484 KDQVSNSAFVERVKARGFEVVYMTEPIDEYVIQHLKEYKGKQLVSVTKEGLELPEDESEK 543 Query: 1664 XXXXXXXXXXXXLCKVMKDILDKKVEKVTVSNRLVSSPCCIVTSQYGWSANMERIMKAQA 1843 LCK+MK ILD KVEKV VSNRLV SPCCIVTSQ+GWSANMERIMKAQA Sbjct: 544 KKREEDKAKFESLCKLMKSILDNKVEKVVVSNRLVDSPCCIVTSQFGWSANMERIMKAQA 603 Query: 1844 LRDSSTMGYMAAKKHLEINPDHSIMKTLKARVDADKNDKSVKDLVMLLYETSLLSSGFTL 2023 LRD++TMGYMA KK LEINPDH I++TL+ + DADKNDK+VKDLV+LL+ETSLLSSGF+L Sbjct: 604 LRDTATMGYMAGKKQLEINPDHPIVETLRQKADADKNDKAVKDLVILLFETSLLSSGFSL 663 Query: 2024 EDPQVHGNRIHRMIKLGLGIEDDD--VEMDATEAGDVPVVASTAEEDAGKMEEVD 2182 + PQVH +RI+RMIKLGLGI++D+ DA AGD P + EDA MEEVD Sbjct: 664 DSPQVHASRIYRMIKLGLGIDEDEPMTTDDAQSAGDAPSLVEDT-EDASHMEEVD 717
>sp|P11501|HS90A_CHICK Heat shock protein HSP 90-alpha Length = 728 Score = 982 bits (2538), Expect = 0.0 Identities = 498/716 (69%), Positives = 570/716 (79%), Gaps = 1/716 (0%) Frame = +2 Query: 38 DADTETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYKSLTDPSVLDTA 217 + + ETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRY+SLTDPS LD+ Sbjct: 13 EEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYESLTDPSKLDSG 72 Query: 218 KELMIKVIPDKDAGTLTIIDSGNGMTKADLINNLGTIARSGTKAFMEALQAGADISMIGQ 397 K+L I +IP+K TLTI+D+G GMTKADL+NNLGTIA+SGTKAFMEALQAGADISMIGQ Sbjct: 73 KDLKINLIPNKHDRTLTIVDTGIGMTKADLVNNLGTIAKSGTKAFMEALQAGADISMIGQ 132 Query: 398 FGVGFYSSYLVADKVQVISKSNDDEQYLWESSAGGSFTIKNDNSEPLGRGTKIILFMKED 577 FGVG YS+YLVA+KV VI+K NDDEQY WESSAGGSFT++ DN EPLGRGTK+IL +KED Sbjct: 133 FGVGSYSAYLVAEKVTVITKHNDDEQYAWESSAGGSFTVRLDNGEPLGRGTKVILHLKED 192 Query: 578 QVEYLEERKIKDIIKKHSQFIGYPIKLVLEKERXXXXXXXXXXXXXXXXXXXXXXXXXXX 757 Q EYLEER+IK+I+KKHSQFIGYPI+L +EKER Sbjct: 193 QTEYLEERRIKEIVKKHSQFIGYPIRLFVEKERDKEVSDDEAEEKEEEKEEKEEKTEDKP 252 Query: 758 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXLNKTKPIWTRNPDDVSQEDYAEFYKSL 937 LNKTKPIWTRNPDD++ E+Y EFYKSL Sbjct: 253 EIEDVGSDEEEEKKDGDKKKKKKIKEKYIDEEELNKTKPIWTRNPDDITNEEYGEFYKSL 312 Query: 938 TNDWEDHLAVKHFSVEGQLEFRALLFCPKRAPFDLFENKKKRNNIKLYVRRVFIMDNCED 1117 TNDWEDHLAVKHFSVEGQLEFRALLF P+RAPFDLFEN+KK+NNIKLYVRRVFIMDNCE+ Sbjct: 313 TNDWEDHLAVKHFSVEGQLEFRALLFVPRRAPFDLFENRKKKNNIKLYVRRVFIMDNCEE 372 Query: 1118 LIPEYLNFVKGVVDSEDLPLNISREMLQQNKILKVIRKNLVKKCIELFEEIMEDKDNYKK 1297 LIPEYLNF++GVVDSEDLPLNISREMLQQ+KILKVIRKNLVKKC+ELF E+ EDK+NYKK Sbjct: 373 LIPEYLNFMRGVVDSEDLPLNISREMLQQSKILKVIRKNLVKKCLELFTELAEDKENYKK 432 Query: 1298 FYEQFSKNIKLGIHEDSVNRNKIAEFLRYYSTSSGDEMCSLKDYVSRMKENQKDIYYITG 1477 FYEQFSKNIKLGIHEDS NR K++E LRYY+++SGDEM SLKDY +RMKENQK +YYITG Sbjct: 433 FYEQFSKNIKLGIHEDSQNRKKLSELLRYYTSASGDEMVSLKDYCTRMKENQKHVYYITG 492 Query: 1478 ESKDNVNHSAFVERLTKRGFEVLLMVDPIDEYSVQQLKEYDGKKLVCVTXXXXXXXXXXX 1657 E+KD V +SAFVERL K G EV+ M++PIDEY VQQLKE++GK LV VT Sbjct: 493 ETKDQVANSAFVERLRKHGLEVIYMIEPIDEYCVQQLKEFEGKTLVSVTKEGLELPEDEE 552 Query: 1658 XXXXXXXXXXXXXXLCKVMKDILDKKVEKVTVSNRLVSSPCCIVTSQYGWSANMERIMKA 1837 LCK+MKDIL+KKVEKV VSNRLV+SPCCIVTS YGW+ANMERIMKA Sbjct: 553 EKKKQEEKKAKFENLCKIMKDILEKKVEKVVVSNRLVTSPCCIVTSTYGWTANMERIMKA 612 Query: 1838 QALRDSSTMGYMAAKKHLEINPDHSIMKTLKARVDADKNDKSVKDLVMLLYETSLLSSGF 2017 QALRD+STMGYMAAKKHLEINPDHSI++TL+ + +ADKNDKSVKDLV+LLYET+LLSSGF Sbjct: 613 QALRDNSTMGYMAAKKHLEINPDHSIIETLRQKAEADKNDKSVKDLVILLYETALLSSGF 672 Query: 2018 TLEDPQVHGNRIHRMIKLGLGIEDDDVEM-DATEAGDVPVVASTAEEDAGKMEEVD 2182 +LEDPQ H NRI+RMIKLGLGI++DD +A+ A + ++D +MEEVD Sbjct: 673 SLEDPQTHANRIYRMIKLGLGIDEDDTAAEEASPAVTEEMPPLEGDDDTSRMEEVD 728
>sp|P08238|HS90B_HUMAN Heat shock protein HSP 90-beta (HSP 84) (HSP 90) Length = 724 Score = 975 bits (2520), Expect = 0.0 Identities = 492/718 (68%), Positives = 571/718 (79%), Gaps = 3/718 (0%) Frame = +2 Query: 38 DADTETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYKSLTDPSVLDTA 217 + + ETFAFQAEIAQLMSLIINTFYSNKEIFLRELISN+SDALDKIRY+SLTDPS LD+ Sbjct: 9 EEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNASDALDKIRYESLTDPSKLDSG 68 Query: 218 KELMIKVIPDKDAGTLTIIDSGNGMTKADLINNLGTIARSGTKAFMEALQAGADISMIGQ 397 KEL I +IP+ TLT++D+G GMTKADLINNLGTIA+SGTKAFMEALQAGADISMIGQ Sbjct: 69 KELKIDIIPNPQERTLTLVDTGIGMTKADLINNLGTIAKSGTKAFMEALQAGADISMIGQ 128 Query: 398 FGVGFYSSYLVADKVQVISKSNDDEQYLWESSAGGSFTIKNDNSEPLGRGTKIILFMKED 577 FGVGFYS+YLVA+KV VI+K NDDEQY WESSAGGSFT++ D+ EP+GRGTK+IL +KED Sbjct: 129 FGVGFYSAYLVAEKVVVITKHNDDEQYAWESSAGGSFTVRADHGEPIGRGTKVILHLKED 188 Query: 578 QVEYLEERKIKDIIKKHSQFIGYPIKLVLEKERXXXXXXXXXXXXXXXXXXXXXXXXXXX 757 Q EYLEER++K+++KKHSQFIGYPI L LEKER Sbjct: 189 QTEYLEERRVKEVVKKHSQFIGYPITLYLEKEREKEISDDEAEEEKGEKEEEDKDDEEKP 248 Query: 758 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXLNKTKPIWTRNPDDVSQEDYAEFYKSL 937 LNKTKPIWTRNPDD++QE+Y EFYKSL Sbjct: 249 KIEDVGSDEEDDSGKDKKKKTKKIKEKYIDQEELNKTKPIWTRNPDDITQEEYGEFYKSL 308 Query: 938 TNDWEDHLAVKHFSVEGQLEFRALLFCPKRAPFDLFENKKKRNNIKLYVRRVFIMDNCED 1117 TNDWEDHLAVKHFSVEGQLEFRALLF P+RAPFDLFENKKK+NNIKLYVRRVFIMD+C++ Sbjct: 309 TNDWEDHLAVKHFSVEGQLEFRALLFIPRRAPFDLFENKKKKNNIKLYVRRVFIMDSCDE 368 Query: 1118 LIPEYLNFVKGVVDSEDLPLNISREMLQQNKILKVIRKNLVKKCIELFEEIMEDKDNYKK 1297 LIPEYLNF++GVVDSEDLPLNISREMLQQ+KILKVIRKN+VKKC+ELF E+ EDK+NYKK Sbjct: 369 LIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNIVKKCLELFSELAEDKENYKK 428 Query: 1298 FYEQFSKNIKLGIHEDSVNRNKIAEFLRYYSTSSGDEMCSLKDYVSRMKENQKDIYYITG 1477 FYE FSKN+KLGIHEDS NR +++E LRY+++ SGDEM SL +YVSRMKE QK IYYITG Sbjct: 429 FYEAFSKNLKLGIHEDSTNRRRLSELLRYHTSQSGDEMTSLSEYVSRMKETQKSIYYITG 488 Query: 1478 ESKDNVNHSAFVERLTKRGFEVLLMVDPIDEYSVQQLKEYDGKKLVCVTXXXXXXXXXXX 1657 ESK+ V +SAFVER+ KRGFEV+ M +PIDEY VQQLKE+DGK LV VT Sbjct: 489 ESKEQVANSAFVERVRKRGFEVVYMTEPIDEYCVQQLKEFDGKSLVSVTKEGLELPEDEE 548 Query: 1658 XXXXXXXXXXXXXXLCKVMKDILDKKVEKVTVSNRLVSSPCCIVTSQYGWSANMERIMKA 1837 LCK+MK+ILDKKVEKVT+SNRLVSSPCCIVTS YGW+ANMERIMKA Sbjct: 549 EKKKMEESKAKFENLCKLMKEILDKKVEKVTISNRLVSSPCCIVTSTYGWTANMERIMKA 608 Query: 1838 QALRDSSTMGYMAAKKHLEINPDHSIMKTLKARVDADKNDKSVKDLVMLLYETSLLSSGF 2017 QALRD+STMGYM AKKHLEINPDH I++TL+ + +ADKNDK+VKDLV+LL+ET+LLSSGF Sbjct: 609 QALRDNSTMGYMMAKKHLEINPDHPIVETLRQKAEADKNDKAVKDLVVLLFETALLSSGF 668 Query: 2018 TLEDPQVHGNRIHRMIKLGLGIEDDDV---EMDATEAGDVPVVASTAEEDAGKMEEVD 2182 +LEDPQ H NRI+RMIKLGLGI++D+V E +A ++P + +EDA +MEEVD Sbjct: 669 SLEDPQTHSNRIYRMIKLGLGIDEDEVAAEEPNAAVPDEIPPL--EGDEDASRMEEVD 724
>sp|Q9GKX8|HS90B_HORSE Heat shock protein HSP 90-beta (HSP 84) Length = 724 Score = 974 bits (2519), Expect = 0.0 Identities = 492/718 (68%), Positives = 570/718 (79%), Gaps = 3/718 (0%) Frame = +2 Query: 38 DADTETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYKSLTDPSVLDTA 217 + + ETFAFQAEIAQLMSLIINTFYSNKEIFLRELISN+SDALDKIRY+SLTDPS LD+ Sbjct: 9 EEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNASDALDKIRYESLTDPSKLDSG 68 Query: 218 KELMIKVIPDKDAGTLTIIDSGNGMTKADLINNLGTIARSGTKAFMEALQAGADISMIGQ 397 KEL I +IP+ TLT++D+G GMTKADLINNLGTIA+SGTKAFMEALQAGADISMIGQ Sbjct: 69 KELKIDIIPNPQERTLTLVDTGIGMTKADLINNLGTIAKSGTKAFMEALQAGADISMIGQ 128 Query: 398 FGVGFYSSYLVADKVQVISKSNDDEQYLWESSAGGSFTIKNDNSEPLGRGTKIILFMKED 577 FGVGFYS+YLVA+KV VI+K NDDEQY WESSAGGSFT++ D+ EP+GRGTK+IL +KED Sbjct: 129 FGVGFYSAYLVAEKVVVITKHNDDEQYAWESSAGGSFTVRADHGEPIGRGTKVILHLKED 188 Query: 578 QVEYLEERKIKDIIKKHSQFIGYPIKLVLEKERXXXXXXXXXXXXXXXXXXXXXXXXXXX 757 Q EYLEER++K+++KKHSQFIGYPI L LEKER Sbjct: 189 QTEYLEERRVKEVVKKHSQFIGYPITLYLEKEREKEISDDEAEEEKGEKEEEDKDDEEKP 248 Query: 758 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXLNKTKPIWTRNPDDVSQEDYAEFYKSL 937 LNKTKPIWTRNPDD++QE+Y EFYKSL Sbjct: 249 KIEDVGSDEEDDSGKDKKKKTKKIKEKYIDQEELNKTKPIWTRNPDDITQEEYGEFYKSL 308 Query: 938 TNDWEDHLAVKHFSVEGQLEFRALLFCPKRAPFDLFENKKKRNNIKLYVRRVFIMDNCED 1117 TNDWEDHLAVKHFSVEGQLEFRALLF P+RAPFDLFENKKK+NNIKLYVRRVFIMD+C++ Sbjct: 309 TNDWEDHLAVKHFSVEGQLEFRALLFIPRRAPFDLFENKKKKNNIKLYVRRVFIMDSCDE 368 Query: 1118 LIPEYLNFVKGVVDSEDLPLNISREMLQQNKILKVIRKNLVKKCIELFEEIMEDKDNYKK 1297 LIPEYLNF++GVVDSEDLPLNISREMLQQ+KILKVIRKN+VKKC+ELF E+ EDK+NYKK Sbjct: 369 LIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNIVKKCLELFSELAEDKENYKK 428 Query: 1298 FYEQFSKNIKLGIHEDSVNRNKIAEFLRYYSTSSGDEMCSLKDYVSRMKENQKDIYYITG 1477 FYE FSKN+KLGIHEDS NR +++E LRY+++ SGDEM SL +YVSRMKE QK IYYITG Sbjct: 429 FYEAFSKNLKLGIHEDSTNRRRLSELLRYHTSQSGDEMTSLSEYVSRMKETQKSIYYITG 488 Query: 1478 ESKDNVNHSAFVERLTKRGFEVLLMVDPIDEYSVQQLKEYDGKKLVCVTXXXXXXXXXXX 1657 ESK+ V +SAFVER+ KRGFEV+ M +PIDEY VQQLKE+DGK LV VT Sbjct: 489 ESKEQVANSAFVERVRKRGFEVVYMTEPIDEYCVQQLKEFDGKSLVSVTKEGLELPEDEE 548 Query: 1658 XXXXXXXXXXXXXXLCKVMKDILDKKVEKVTVSNRLVSSPCCIVTSQYGWSANMERIMKA 1837 LCK+MK+ILDKKVEKVT+SNRLVSSPCCIVTS YGW+ANMERIMKA Sbjct: 549 EKKKMEESKAKFENLCKLMKEILDKKVEKVTISNRLVSSPCCIVTSTYGWTANMERIMKA 608 Query: 1838 QALRDSSTMGYMAAKKHLEINPDHSIMKTLKARVDADKNDKSVKDLVMLLYETSLLSSGF 2017 QALRD+STMGYM AKKHLEINPDH I++TL+ + +ADKNDK+VKDLV+LL+ET+LLSSGF Sbjct: 609 QALRDNSTMGYMMAKKHLEINPDHPIVETLRQKAEADKNDKAVKDLVVLLFETALLSSGF 668 Query: 2018 TLEDPQVHGNRIHRMIKLGLGIEDDDV---EMDATEAGDVPVVASTAEEDAGKMEEVD 2182 +LEDPQ H NRI+RMIKLGLGI++D+V E A ++P + +EDA +MEEVD Sbjct: 669 SLEDPQTHSNRIYRMIKLGLGIDEDEVAAEEPSAAVPDEIPPL--EGDEDASRMEEVD 724
>sp|O57521|HS90B_BRARE Heat shock protein HSP 90-beta Length = 725 Score = 971 bits (2511), Expect = 0.0 Identities = 491/718 (68%), Positives = 564/718 (78%), Gaps = 3/718 (0%) Frame = +2 Query: 38 DADTETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYKSLTDPSVLDTA 217 + + ETFAFQAEIAQLMSLIINTFYSNKEIFLREL+SN+SDALDKIRY+SLTDP+ LD+ Sbjct: 8 EEEAETFAFQAEIAQLMSLIINTFYSNKEIFLRELVSNASDALDKIRYESLTDPTKLDSG 67 Query: 218 KELMIKVIPDKDAGTLTIIDSGNGMTKADLINNLGTIARSGTKAFMEALQAGADISMIGQ 397 K+L I +IP+ TLT+ID+G GMTKADLINNLGTIA+SGTKAFMEALQAGADISMIGQ Sbjct: 68 KDLKIDIIPNVQERTLTLIDTGIGMTKADLINNLGTIAKSGTKAFMEALQAGADISMIGQ 127 Query: 398 FGVGFYSSYLVADKVQVISKSNDDEQYLWESSAGGSFTIKNDNSEPLGRGTKIILFMKED 577 FGVGFYS+YLVA+KV VI+K NDDEQY WESSAGGSFT+K D+ EP+GRGTK+IL +KED Sbjct: 128 FGVGFYSAYLVAEKVTVITKHNDDEQYAWESSAGGSFTVKVDHGEPIGRGTKVILHLKED 187 Query: 578 QVEYLEERKIKDIIKKHSQFIGYPIKLVLEKERXXXXXXXXXXXXXXXXXXXXXXXXXXX 757 Q EY+EE+++K+++KKHSQFIGYPI L +EKER Sbjct: 188 QTEYIEEKRVKEVVKKHSQFIGYPITLYVEKERDKEISDDEAEEEKAEKEEKEEEGEDKP 247 Query: 758 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXLNKTKPIWTRNPDDVSQEDYAEFYKSL 937 LNKTKPIWTRNPDD+S E+Y EFYKSL Sbjct: 248 KIEDVGSDDEEDTKDKDKKKKKKIKEKYIDQEELNKTKPIWTRNPDDISNEEYGEFYKSL 307 Query: 938 TNDWEDHLAVKHFSVEGQLEFRALLFCPKRAPFDLFENKKKRNNIKLYVRRVFIMDNCED 1117 TNDWEDHLAVKHFSVEGQLEFRALLF P+RAPFDLFENKKK+NNIKLYVRRVFIMDNCE+ Sbjct: 308 TNDWEDHLAVKHFSVEGQLEFRALLFIPRRAPFDLFENKKKKNNIKLYVRRVFIMDNCEE 367 Query: 1118 LIPEYLNFVKGVVDSEDLPLNISREMLQQNKILKVIRKNLVKKCIELFEEIMEDKDNYKK 1297 LIPEYLNF++GVVDSEDLPLNISREMLQQ+KILKVIRKN+VKKC+ELF E+ EDKDNYKK Sbjct: 368 LIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNIVKKCLELFAELAEDKDNYKK 427 Query: 1298 FYEQFSKNIKLGIHEDSVNRNKIAEFLRYYSTSSGDEMCSLKDYVSRMKENQKDIYYITG 1477 FY+ FSKN+KLGIHED NR K++E LRY S+ SGDEM SL +YVSRMKENQK IYYITG Sbjct: 428 FYDAFSKNLKLGIHEDCQNRKKLSELLRYQSSQSGDEMTSLTEYVSRMKENQKSIYYITG 487 Query: 1478 ESKDNVNHSAFVERLTKRGFEVLLMVDPIDEYSVQQLKEYDGKKLVCVTXXXXXXXXXXX 1657 ESKD V HSAFVER+ KRGFEVL M +PIDEY VQQLK++DGK LV VT Sbjct: 488 ESKDQVAHSAFVERVCKRGFEVLYMTEPIDEYCVQQLKDFDGKSLVSVTKEGLELPEDED 547 Query: 1658 XXXXXXXXXXXXXXLCKVMKDILDKKVEKVTVSNRLVSSPCCIVTSQYGWSANMERIMKA 1837 LCK+MK+ILDKKVEKVTVSNRLVSSPCCIVTS YGW+ANMERIMKA Sbjct: 548 EKKKMEEDKAKFENLCKLMKEILDKKVEKVTVSNRLVSSPCCIVTSTYGWTANMERIMKA 607 Query: 1838 QALRDSSTMGYMAAKKHLEINPDHSIMKTLKARVDADKNDKSVKDLVMLLYETSLLSSGF 2017 QALRD+STMGYM A KHLEINPDH IM+TL+ + +ADKN K+VKDLV+LL+ET+LLSSGF Sbjct: 608 QALRDNSTMGYMMANKHLEINPDHPIMETLRQKAEADKNTKAVKDLVILLFETALLSSGF 667 Query: 2018 TLEDPQVHGNRIHRMIKLGLGIEDDD---VEMDATEAGDVPVVASTAEEDAGKMEEVD 2182 +L+DPQ H NRI+RMIKLGLGI++D+ VE + A + ++DA +MEEVD Sbjct: 668 SLDDPQTHSNRIYRMIKLGLGIDEDEDVPVEEPTSAAAPEDIPPLEGDDDASRMEEVD 725
>sp|P11499|HS90B_MOUSE Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA) Length = 724 Score = 970 bits (2508), Expect = 0.0 Identities = 489/718 (68%), Positives = 568/718 (79%), Gaps = 3/718 (0%) Frame = +2 Query: 38 DADTETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYKSLTDPSVLDTA 217 + + ETFAFQAEIAQLMSLIINTFYSNKEIFLRELISN+SDALDKIRY+SLTDPS LD+ Sbjct: 9 EEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNASDALDKIRYESLTDPSKLDSG 68 Query: 218 KELMIKVIPDKDAGTLTIIDSGNGMTKADLINNLGTIARSGTKAFMEALQAGADISMIGQ 397 KEL I ++P+ TLT++D+G GMTKADLINNLGTIA+SGTKAFMEALQAGADISMIGQ Sbjct: 69 KELKIDILPNPQERTLTLVDTGIGMTKADLINNLGTIAKSGTKAFMEALQAGADISMIGQ 128 Query: 398 FGVGFYSSYLVADKVQVISKSNDDEQYLWESSAGGSFTIKNDNSEPLGRGTKIILFMKED 577 FGVGFYS+YLVA+KV VI+K NDDEQY WESSAGGSFT++ D+ EP+GRGTK+IL +KED Sbjct: 129 FGVGFYSAYLVAEKVVVITKHNDDEQYAWESSAGGSFTVRADHGEPIGRGTKVILHLKED 188 Query: 578 QVEYLEERKIKDIIKKHSQFIGYPIKLVLEKERXXXXXXXXXXXXXXXXXXXXXXXXXXX 757 Q EYLEER++K+++KKHSQFIGYPI L LEKER Sbjct: 189 QTEYLEERRVKEVVKKHSQFIGYPITLYLEKEREKEISDDEAEEEKGEKEEEDKEDEEKP 248 Query: 758 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXLNKTKPIWTRNPDDVSQEDYAEFYKSL 937 LNKTKPIWTRNPDD++QE+Y EFYKSL Sbjct: 249 KIEDVGSDEEDDSGKDKKKKTKKIKEKYIDQEELNKTKPIWTRNPDDITQEEYGEFYKSL 308 Query: 938 TNDWEDHLAVKHFSVEGQLEFRALLFCPKRAPFDLFENKKKRNNIKLYVRRVFIMDNCED 1117 TNDWEDHLAVKHFSVEGQLEFRA LF P+RAPFDLFENKKK+NNIKLYVRRVFIMD+C++ Sbjct: 309 TNDWEDHLAVKHFSVEGQLEFRAFLFIPRRAPFDLFENKKKKNNIKLYVRRVFIMDSCDE 368 Query: 1118 LIPEYLNFVKGVVDSEDLPLNISREMLQQNKILKVIRKNLVKKCIELFEEIMEDKDNYKK 1297 LIPEYLNF++GVVDSEDLPLNISREMLQQ+KILKVIRKN+VKKC+ELF E+ EDK+NYKK Sbjct: 369 LIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNIVKKCLELFSELAEDKENYKK 428 Query: 1298 FYEQFSKNIKLGIHEDSVNRNKIAEFLRYYSTSSGDEMCSLKDYVSRMKENQKDIYYITG 1477 FYE FSKN+KLGIHEDS NR +++E LRY+++ SGDEM SL +YVSRMKE QK IYYITG Sbjct: 429 FYEAFSKNLKLGIHEDSTNRRRLSELLRYHTSQSGDEMTSLSEYVSRMKETQKSIYYITG 488 Query: 1478 ESKDNVNHSAFVERLTKRGFEVLLMVDPIDEYSVQQLKEYDGKKLVCVTXXXXXXXXXXX 1657 ESK+ V + AFVER+ KRGFEV+ M +PIDEY VQQLKE+DGK LV VT Sbjct: 489 ESKEQVANPAFVERVRKRGFEVVYMTEPIDEYCVQQLKEFDGKSLVSVTKEGLELPEDEE 548 Query: 1658 XXXXXXXXXXXXXXLCKVMKDILDKKVEKVTVSNRLVSSPCCIVTSQYGWSANMERIMKA 1837 LCK+MK+ILDKKVEKVT+SNRLVSSPCCIVTS YGW+ANMERIMKA Sbjct: 549 EKKKMEESKAKFENLCKLMKEILDKKVEKVTISNRLVSSPCCIVTSTYGWTANMERIMKA 608 Query: 1838 QALRDSSTMGYMAAKKHLEINPDHSIMKTLKARVDADKNDKSVKDLVMLLYETSLLSSGF 2017 QALRD+STMGYM AKKHLEINPDH I++TL+ + +ADKNDK+VKDLV+LL+ET+LLSSGF Sbjct: 609 QALRDNSTMGYMMAKKHLEINPDHPIVETLRQKAEADKNDKAVKDLVVLLFETALLSSGF 668 Query: 2018 TLEDPQVHGNRIHRMIKLGLGIEDDDV---EMDATEAGDVPVVASTAEEDAGKMEEVD 2182 +LEDPQ H NRI+RMIKLGLGI++D+V E A ++P + +EDA +MEEVD Sbjct: 669 SLEDPQTHSNRIYRMIKLGLGIDEDEVTAEEPSAAVPDEIPPL--EGDEDASRMEEVD 724
>sp|P34058|HS90B_RAT Heat shock protein HSP 90-beta (HSP 84) Length = 724 Score = 957 bits (2475), Expect = 0.0 Identities = 485/718 (67%), Positives = 562/718 (78%), Gaps = 3/718 (0%) Frame = +2 Query: 38 DADTETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYKSLTDPSVLDTA 217 + + ETFAFQAEIAQLMSLIINTFYSNKEIFLRELISN+SDALDKIRY+SLTDPS LD+ Sbjct: 9 EEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNASDALDKIRYESLTDPSKLDSG 68 Query: 218 KELMIKVIPDKDAGTLTIIDSGNGMTKADLINNLGTIARSGTKAFMEALQAGADISMIGQ 397 KEL I +IP+ TLT++D+G GMTKADLINNLGTIA+SGTKAFMEALQAGADISMIGQ Sbjct: 69 KELKIDIIPNPQEATLTLVDTGIGMTKADLINNLGTIAKSGTKAFMEALQAGADISMIGQ 128 Query: 398 FGVGFYSSYLVADKVQVISKSNDDEQYLWESSAGGSFTIKNDNSEPLGRGTKIILFMKED 577 FGVGFYS+YLVA+KV VI+K NDDEQY WESSAGGSFT++ D+ EP+GRGTK+IL +KED Sbjct: 129 FGVGFYSAYLVAEKVVVITKHNDDEQYAWESSAGGSFTVRADHGEPIGRGTKVILHLKED 188 Query: 578 QVEYLEERKIKDIIKKHSQFIGYPIKLVLEKERXXXXXXXXXXXXXXXXXXXXXXXXXXX 757 Q EYLEER++K+++KKHSQFIGYPI L LEKER Sbjct: 189 QTEYLEERRVKEVVKKHSQFIGYPITLYLEKEREKEISDDEAEEEKGEKEEEDKEDEEKP 248 Query: 758 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXLNKTKPIWTRNPDDVSQEDYAEFYKSL 937 LNKTKPIWTRNPDD++QE+Y EFYKSL Sbjct: 249 KIEDVGSDEEDDSGKDKKKKTKKIKEKYIDQEELNKTKPIWTRNPDDITQEEYGEFYKSL 308 Query: 938 TNDWEDHLAVKHFSVEGQLEFRALLFCPKRAPFDLFENKKKRNNIKLYVRRVFIMDNCED 1117 TNDWEDHLAVKHFSVEGQLEFRALLF P+RAPFDLFENKKK+NNIKLYVRRVFIMD+C+D Sbjct: 309 TNDWEDHLAVKHFSVEGQLEFRALLFIPRRAPFDLFENKKKKNNIKLYVRRVFIMDSCDD 368 Query: 1118 LIPEYLNFVKGVVDSEDLPLNISREMLQQNKILKVIRKNLVKKCIELFEEIMEDKDNYKK 1297 LIPEYLNF++GVVDSEDLPLNISREMLQQ+KILKVIRKN+VKKC+ELF E+ EDK+NYKK Sbjct: 369 LIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNIVKKCLELFSELAEDKENYKK 428 Query: 1298 FYEQFSKNIKLGIHEDSVNRNKIAEFLRYYSTSSGDEMCSLKDYVSRMKENQKDIYYITG 1477 FYE FSKN+KLGIHEDS NR +++E LRY+++ SGDEM SL +YVSRMKE QK IYYITG Sbjct: 429 FYEAFSKNLKLGIHEDSTNRRRLSELLRYHTSQSGDEMTSLSEYVSRMKETQKSIYYITG 488 Query: 1478 ESKDNVNHSAFVERLTKRGFEVLLMVDPIDEYSVQQLKEYDGKKLVCVTXXXXXXXXXXX 1657 ESK+ V +SAFVER+ KRGFEV+ M +PIDEY VQQLKE+DGK LV VT Sbjct: 489 ESKEQVANSAFVERVRKRGFEVVYMTEPIDEYCVQQLKEFDGKSLVSVTKEGLELPEDEE 548 Query: 1658 XXXXXXXXXXXXXXLCKVMKDILDKKVEKVTVSNRLVSSPCCIVTSQYGWSANMERIMKA 1837 LCK+MK+ILDKKVEKVT+SNRLVSSPCCIVTS YGW+ANMERIMKA Sbjct: 549 EKKKMEESKARFENLCKLMKEILDKKVEKVTISNRLVSSPCCIVTSTYGWTANMERIMKA 608 Query: 1838 QALRDSSTMGYMAAKKHLEINPDHSIMKTLKARVDADKNDKSVKDLVMLLYETSLLSSGF 2017 QALRD+STMGYM AKKHLEINPDH I++TL+ + +ADKNDK+VKDLV+LL+ET+L S Sbjct: 609 QALRDNSTMGYMMAKKHLEINPDHPIVETLRQKAEADKNDKAVKDLVVLLFETALSSLAS 668 Query: 2018 TLEDPQVHGNRIHRMIKLGLGIEDDDV---EMDATEAGDVPVVASTAEEDAGKMEEVD 2182 P+ H NRI+RMIKLGLGI++D+V E A ++P + +EDA +MEEVD Sbjct: 669 HFRRPKTHSNRIYRMIKLGLGIDEDEVTAEEPSAAVPDEIPPL--EGDEDASRMEEVD 724
>sp|O61998|HSP90_BRUPA Heat shock protein 90 Length = 717 Score = 671 bits (1730), Expect = 0.0 Identities = 337/447 (75%), Positives = 385/447 (86%), Gaps = 5/447 (1%) Frame = +2 Query: 857 LNKTKPIWTRNPDDVSQEDYAEFYKSLTNDWEDHLAVKHFSVEGQLEFRALLFCPKRAPF 1036 LNKTKPIWTRNPDD+S E+YAEFYKSL+NDWEDHLAVKHFSVEGQLEFRALLF P+RAPF Sbjct: 272 LNKTKPIWTRNPDDISNEEYAEFYKSLSNDWEDHLAVKHFSVEGQLEFRALLFVPQRAPF 331 Query: 1037 DLFENKKKRNNIKLYVRRVFIMDNCEDLIPEYLNFVKGVVDSEDLPLNISREMLQQNKIL 1216 DLFENKK +N IKLYVRRVFIM+NC++L+PEYLNF+KGVVDSEDLPLNISREMLQQ+KIL Sbjct: 332 DLFENKKTKNAIKLYVRRVFIMENCDELMPEYLNFIKGVVDSEDLPLNISREMLQQSKIL 391 Query: 1217 KVIRKNLVKKCIELFEEIMEDKDNYKKFYEQFSKNIKLGIHEDSVNRNKIAEFLRYYSTS 1396 KVIRKNLVKKC+ELF+EI EDKDN+KKFYEQFSKNIKLGIHEDS NR K++EFLR+Y+++ Sbjct: 392 KVIRKNLVKKCLELFDEIAEDKDNFKKFYEQFSKNIKLGIHEDSTNRKKLSEFLRFYTSA 451 Query: 1397 SGDEMCSLKDYVSRMKENQKDIYYITGESKDNVNHSAFVERLTKRGFEVLLMVDPIDEYS 1576 S +EM SLKDYVSRMKENQK IY+ITGES++ V SAFVER+ +RGFEV+ M DPIDEY Sbjct: 452 SSEEMTSLKDYVSRMKENQKQIYFITGESREAVASSAFVERVKRRGFEVIYMTDPIDEYC 511 Query: 1577 VQQLKEYDGKKLVCVTXXXXXXXXXXXXXXXXXXXXXXXXXLCKVMKDILDKKVEKVTVS 1756 VQQLKEYDGKKLV VT LCKVMKDIL+KKVEKV VS Sbjct: 512 VQQLKEYDGKKLVSVTKEGLELPESEEEKKKFEEDKVKFENLCKVMKDILEKKVEKVAVS 571 Query: 1757 NRLVSSPCCIVTSQYGWSANMERIMKAQALRDSSTMGYMAAKKHLEINPDHSIMKTLKAR 1936 NRLVSSPCCIVTS+YGWSANMERIMKAQALRDSSTMGYMAAKKHLEINPDHS++K L+ R Sbjct: 572 NRLVSSPCCIVTSEYGWSANMERIMKAQALRDSSTMGYMAAKKHLEINPDHSVIKALRER 631 Query: 1937 VDADKNDKSVKDLVMLLYETSLLSSGFTLEDPQVHGNRIHRMIKLGLGI-EDDDVEMDAT 2113 V+ADKNDK+VKDLV+LL+ET+LLSSGF+LEDPQ+H +RI+RMIKLGL I ED++ E A+ Sbjct: 632 VEADKNDKTVKDLVVLLFETALLSSGFSLEDPQLHASRIYRMIKLGLDITEDEEEEAIAS 691 Query: 2114 EAGD----VPVVASTAEEDAGKMEEVD 2182 +G+ VP + AEEDA +MEEVD Sbjct: 692 VSGEKDECVPNLVG-AEEDASRMEEVD 717
Score = 324 bits (830), Expect = 8e-88
Identities = 164/209 (78%), Positives = 186/209 (88%)
Frame = +2
Query: 50 ETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYKSLTDPSVLDTAKELM 229
ETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRY++LT+P+ L+T KEL
Sbjct: 8 ETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYQALTEPAELETGKELY 67
Query: 230 IKVIPDKDAGTLTIIDSGNGMTKADLINNLGTIARSGTKAFMEALQAGADISMIGQFGVG 409
IK+ P+K TLTI+D+G GMTKADL+NNLGTIA+SGTKAFMEALQAGADISMIGQFGVG
Sbjct: 68 IKITPNKADKTLTIMDTGIGMTKADLVNNLGTIAKSGTKAFMEALQAGADISMIGQFGVG 127
Query: 410 FYSSYLVADKVQVISKSNDDEQYLWESSAGGSFTIKNDNSEPLGRGTKIILFMKEDQVEY 589
FYS++LVADKV V SK NDD+ Y WESSAGGSF I+ N L RGTKI L++KEDQ +Y
Sbjct: 128 FYSAFLVADKVVVASKHNDDDCYQWESSAGGSFIIRQVNDPELTRGTKITLYIKEDQTDY 187
Query: 590 LEERKIKDIIKKHSQFIGYPIKLVLEKER 676
LEER+IK+I+KKHSQFIGYPIKL +EKER
Sbjct: 188 LEERRIKEIVKKHSQFIGYPIKLTVEKER 216
>sp|P07900|HS90A_HUMAN Heat shock protein HSP 90-alpha (HSP 86) Length = 732 Score = 669 bits (1726), Expect = 0.0 Identities = 330/445 (74%), Positives = 378/445 (84%), Gaps = 3/445 (0%) Frame = +2 Query: 857 LNKTKPIWTRNPDDVSQEDYAEFYKSLTNDWEDHLAVKHFSVEGQLEFRALLFCPKRAPF 1036 LNKTKPIWTRNPDD++ E+Y EFYKSLTNDWEDHLAVKHFSVEGQLEFRALLF P+RAPF Sbjct: 290 LNKTKPIWTRNPDDITNEEYGEFYKSLTNDWEDHLAVKHFSVEGQLEFRALLFVPRRAPF 349 Query: 1037 DLFENKKKRNNIKLYVRRVFIMDNCEDLIPEYLNFVKGVVDSEDLPLNISREMLQQNKIL 1216 DLFEN+KK+NNIKLYVRRVFIMDNCE+LIPEYLNF++GVVDSEDLPLNISREMLQQ+KIL Sbjct: 350 DLFENRKKKNNIKLYVRRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSKIL 409 Query: 1217 KVIRKNLVKKCIELFEEIMEDKDNYKKFYEQFSKNIKLGIHEDSVNRNKIAEFLRYYSTS 1396 KVIRKNLVKKC+ELF E+ EDK+NYKKFYEQFSKNIKLGIHEDS NR K++E LRYY+++ Sbjct: 410 KVIRKNLVKKCLELFTELAEDKENYKKFYEQFSKNIKLGIHEDSQNRKKLSELLRYYTSA 469 Query: 1397 SGDEMCSLKDYVSRMKENQKDIYYITGESKDNVNHSAFVERLTKRGFEVLLMVDPIDEYS 1576 SGDEM SLKDY +RMKENQK IYYITGE+KD V +SAFVERL K G EV+ M++PIDEY Sbjct: 470 SGDEMVSLKDYCTRMKENQKHIYYITGETKDQVANSAFVERLRKHGLEVIYMIEPIDEYC 529 Query: 1577 VQQLKEYDGKKLVCVTXXXXXXXXXXXXXXXXXXXXXXXXXLCKVMKDILDKKVEKVTVS 1756 VQQLKE++GK LV VT LCK+MKDIL+KKVEKV VS Sbjct: 530 VQQLKEFEGKTLVSVTKEGLELPEDEEEKKKQEEKKTKFENLCKIMKDILEKKVEKVVVS 589 Query: 1757 NRLVSSPCCIVTSQYGWSANMERIMKAQALRDSSTMGYMAAKKHLEINPDHSIMKTLKAR 1936 NRLV+SPCCIVTS YGW+ANMERIMKAQALRD+STMGYMAAKKHLEINPDHSI++TL+ + Sbjct: 590 NRLVTSPCCIVTSTYGWTANMERIMKAQALRDNSTMGYMAAKKHLEINPDHSIIETLRQK 649 Query: 1937 VDADKNDKSVKDLVMLLYETSLLSSGFTLEDPQVHGNRIHRMIKLGLGIEDDDVEMDATE 2116 +ADKNDKSVKDLV+LLYET+LLSSGF+LEDPQ H NRI+RMIKLGLGI++DD D T Sbjct: 650 AEADKNDKSVKDLVILLYETALLSSGFSLEDPQTHANRIYRMIKLGLGIDEDDPTADDTS 709 Query: 2117 AG---DVPVVASTAEEDAGKMEEVD 2182 A ++P + ++D +MEEVD Sbjct: 710 AAVTEEMPPL--EGDDDTSRMEEVD 732
Score = 346 bits (887), Expect = 2e-94
Identities = 172/215 (80%), Positives = 194/215 (90%)
Frame = +2
Query: 32 MSDADTETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYKSLTDPSVLD 211
M + + ETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRY++LTDPS LD
Sbjct: 12 MEEEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYETLTDPSKLD 71
Query: 212 TAKELMIKVIPDKDAGTLTIIDSGNGMTKADLINNLGTIARSGTKAFMEALQAGADISMI 391
+ KEL I +IP+K TLTI+D+G GMTKADLINNLGTIA+SGTKAFMEALQAGADISMI
Sbjct: 72 SGKELHINLIPNKQDRTLTIVDTGIGMTKADLINNLGTIAKSGTKAFMEALQAGADISMI 131
Query: 392 GQFGVGFYSSYLVADKVQVISKSNDDEQYLWESSAGGSFTIKNDNSEPLGRGTKIILFMK 571
GQFGVGFYS+YLVA+KV VI+K NDDEQY WESSAGGSFT++ D EP+GRGTK+IL +K
Sbjct: 132 GQFGVGFYSAYLVAEKVTVITKHNDDEQYAWESSAGGSFTVRTDTGEPMGRGTKVILHLK 191
Query: 572 EDQVEYLEERKIKDIIKKHSQFIGYPIKLVLEKER 676
EDQ EYLEER+IK+I+KKHSQFIGYPI L +EKER
Sbjct: 192 EDQTEYLEERRIKEIVKKHSQFIGYPITLFVEKER 226
>sp|O02705|HS90A_PIG Heat shock protein HSP 90-alpha (HSP 86) Length = 733 Score = 667 bits (1722), Expect = 0.0 Identities = 329/445 (73%), Positives = 378/445 (84%), Gaps = 3/445 (0%) Frame = +2 Query: 857 LNKTKPIWTRNPDDVSQEDYAEFYKSLTNDWEDHLAVKHFSVEGQLEFRALLFCPKRAPF 1036 LNKTKPIWTRNPDD++ E+Y EFYKSLTNDWEDHLAVKHFSVEGQLEFRALLF P+RAPF Sbjct: 291 LNKTKPIWTRNPDDITNEEYGEFYKSLTNDWEDHLAVKHFSVEGQLEFRALLFVPRRAPF 350 Query: 1037 DLFENKKKRNNIKLYVRRVFIMDNCEDLIPEYLNFVKGVVDSEDLPLNISREMLQQNKIL 1216 DLFEN+KK+NNIKLYVRRVFIMDNCE+LIPEYLNF++GVVDSEDLPLNISREMLQQ+KIL Sbjct: 351 DLFENRKKKNNIKLYVRRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSKIL 410 Query: 1217 KVIRKNLVKKCIELFEEIMEDKDNYKKFYEQFSKNIKLGIHEDSVNRNKIAEFLRYYSTS 1396 KVIRKNLVKKC+ELF E+ EDK+NYKKFYEQFSKNIKLGIHEDS NR K++E LRYY+++ Sbjct: 411 KVIRKNLVKKCLELFTELAEDKENYKKFYEQFSKNIKLGIHEDSQNRKKLSELLRYYTSA 470 Query: 1397 SGDEMCSLKDYVSRMKENQKDIYYITGESKDNVNHSAFVERLTKRGFEVLLMVDPIDEYS 1576 SGDEM SLKDY +RMKENQK IYYITGE+KD V +SAFVERL K G EV+ M++PIDEY Sbjct: 471 SGDEMVSLKDYCTRMKENQKHIYYITGETKDQVANSAFVERLRKHGLEVIYMIEPIDEYC 530 Query: 1577 VQQLKEYDGKKLVCVTXXXXXXXXXXXXXXXXXXXXXXXXXLCKVMKDILDKKVEKVTVS 1756 VQQLKE++GK LV VT LCK+MKDIL+KKVEKV VS Sbjct: 531 VQQLKEFEGKTLVSVTKEGLELPEDEEEKKKQEEKKTKFENLCKIMKDILEKKVEKVVVS 590 Query: 1757 NRLVSSPCCIVTSQYGWSANMERIMKAQALRDSSTMGYMAAKKHLEINPDHSIMKTLKAR 1936 NRLV+SPCCIVTS YGW+ANMERIMKAQALRD+STMGYMAAKKHLEINPDHSI++TL+ + Sbjct: 591 NRLVTSPCCIVTSTYGWTANMERIMKAQALRDNSTMGYMAAKKHLEINPDHSIIETLRQK 650 Query: 1937 VDADKNDKSVKDLVMLLYETSLLSSGFTLEDPQVHGNRIHRMIKLGLGIEDDDVEMDATE 2116 +ADKNDKSVKDLV+LLYET+LLSSGF+LEDPQ H NRI+RMIKLGLGI++DD D + Sbjct: 651 AEADKNDKSVKDLVILLYETALLSSGFSLEDPQTHANRIYRMIKLGLGIDEDDPTADDSS 710 Query: 2117 AG---DVPVVASTAEEDAGKMEEVD 2182 A ++P + ++D +MEEVD Sbjct: 711 AAVTEEMPPL--EGDDDTSRMEEVD 733
Score = 347 bits (890), Expect = 9e-95
Identities = 173/215 (80%), Positives = 194/215 (90%)
Frame = +2
Query: 32 MSDADTETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYKSLTDPSVLD 211
M + + ETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRY+SLTDPS LD
Sbjct: 12 MEEEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYESLTDPSKLD 71
Query: 212 TAKELMIKVIPDKDAGTLTIIDSGNGMTKADLINNLGTIARSGTKAFMEALQAGADISMI 391
+ KEL I +IP+K TLTI+D+G GMTKADLINNLGTIA+SGTKAFMEALQAGADISMI
Sbjct: 72 SGKELHINLIPNKQDRTLTIVDTGIGMTKADLINNLGTIAKSGTKAFMEALQAGADISMI 131
Query: 392 GQFGVGFYSSYLVADKVQVISKSNDDEQYLWESSAGGSFTIKNDNSEPLGRGTKIILFMK 571
GQFGVGFYS+YLVA+KV VI+K NDDEQY WESSAGGSFT++ D EP+GRGTK+IL +K
Sbjct: 132 GQFGVGFYSAYLVAEKVTVITKHNDDEQYAWESSAGGSFTVRTDTGEPMGRGTKVILHLK 191
Query: 572 EDQVEYLEERKIKDIIKKHSQFIGYPIKLVLEKER 676
EDQ EYLEER+IK+I+KKHSQFIGYPI L +EKER
Sbjct: 192 EDQTEYLEERRIKEIVKKHSQFIGYPITLFVEKER 226
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 206,767,595
Number of Sequences: 369166
Number of extensions: 3904166
Number of successful extensions: 12783
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 11294
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 12249
length of database: 68,354,980
effective HSP length: 118
effective length of database: 46,556,250
effective search space used: 30075337500
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail
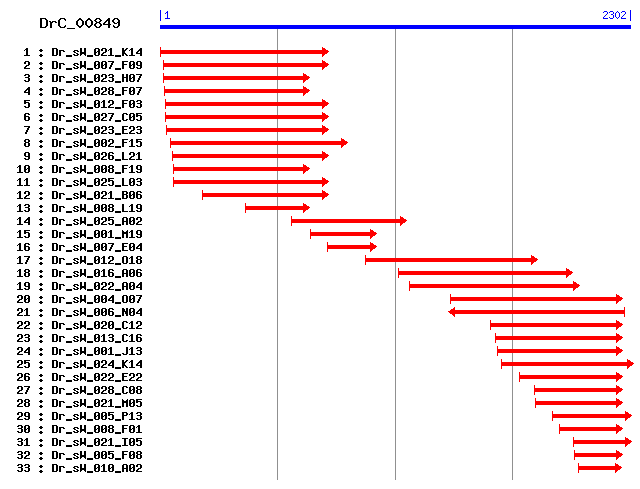
- Dr_sW_023_H07
- Dr_sW_008_L19
- Dr_sW_008_F19
- Dr_sW_028_F07
- Dr_sW_021_K14
- Dr_sW_007_F09
- Dr_sW_023_E23
- Dr_sW_012_F03
- Dr_sW_025_L03
- Dr_sW_027_C05
- Dr_sW_021_B06
- Dr_sW_026_L21
- Dr_sW_002_F15
- Dr_sW_001_M19
- Dr_sW_007_E04
- Dr_sW_025_A02
- Dr_sW_012_O18
- Dr_sW_016_A06
- Dr_sW_022_A04
- Dr_sW_004_O07
- Dr_sW_010_A02
- Dr_sW_005_F08
- Dr_sW_008_F01
- Dr_sW_021_M05
- Dr_sW_028_C08
- Dr_sW_022_E22
- Dr_sW_013_C16
- Dr_sW_020_C12
- Dr_sW_006_N04
- Dr_sW_001_J13
- Dr_sW_024_K14
- Dr_sW_005_P13
- Dr_sW_021_I05