DrC_00422
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00422 (1391 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P41383|TBA2_PATVU Tubulin alpha-2/alpha-4 chain 760 0.0 sp|P18258|TBA1_PARLI Tubulin alpha-1 chain 759 0.0 sp|Q91060|TBA_NOTVI Tubulin alpha chain 759 0.0 sp|P06603|TBA1_DROME Tubulin alpha-1 chain 758 0.0 sp|P68369|TBA1_MOUSE Tubulin alpha-1 chain (Alpha-tubulin 1... 757 0.0 sp|Q8T6A5|TBA1_APLCA Tubulin alpha-1 chain 757 0.0 sp|P05214|TBA3_MOUSE Tubulin alpha-3/alpha-7 chain (Alpha-t... 756 0.0 sp|P68361|TBA1_CRIGR Tubulin alpha-1 chain (Alpha-tubulin 1... 755 0.0 sp|P06605|TBA3_DROME Tubulin alpha-3 chain 754 0.0 sp|P52273|TBA_BOMMO Tubulin alpha chain 754 0.0
>sp|P41383|TBA2_PATVU Tubulin alpha-2/alpha-4 chain Length = 452 Score = 760 bits (1963), Expect = 0.0 Identities = 364/433 (84%), Positives = 392/433 (90%) Frame = +3 Query: 3 ISIHIGQAGVQMGNACWELYCLEHGIQQDGQMPSEKPKHGNEDCFNTFFSETGNGKYVPR 182 IS+HIGQAGVQMGNACWELYCLEHGIQ DGQMPS+K G +D FNTFFSETG GK+VPR Sbjct: 5 ISVHIGQAGVQMGNACWELYCLEHGIQPDGQMPSDKTIGGGDDSFNTFFSETGAGKHVPR 64 Query: 183 ALFVDLEPSVIDEVRNGAYRQLFHPEQLISGKEDAANNYARGHYTVGKELIDQVLDRIRK 362 A+FVDLEP+V+DEVR G YRQLFHPEQLI+GKEDAANNYARGHYTVGKE+ID VLDRIRK Sbjct: 65 AVFVDLEPTVVDEVRTGTYRQLFHPEQLITGKEDAANNYARGHYTVGKEIIDLVLDRIRK 124 Query: 363 VADNCTGLQGFLMFHXXXXXXXXXXXXLLMERLSVDYGKKSKLEFAVYPAPQIATAVVEP 542 +AD CTGLQGFL+FH LLMERLSVDYGKKSKLEFA+YPAPQI+TAVVEP Sbjct: 125 LADQCTGLQGFLIFHSFGGGTGSGFSSLLMERLSVDYGKKSKLEFAIYPAPQISTAVVEP 184 Query: 543 YNSILTTHTTLEHSDCAFMVDNEAIYDICKKNLGIERPTYTSLNRLIGQIVSSITASLRF 722 YNSILTTHTTLEHSDCAFMVDNEAIYDIC++NL IERPTYT+LNRLIGQIVSSITASLRF Sbjct: 185 YNSILTTHTTLEHSDCAFMVDNEAIYDICRRNLDIERPTYTNLNRLIGQIVSSITASLRF 244 Query: 723 DGALNVDLNEFQTNLVPYPRIHFPLATYAPIICADRAFHEQLSVAEITNSCFEHNNQMVK 902 DGALNVDL EFQTNLVPYPRIHFPLATYAP+I A++A+HEQL+VAEITN+CFE NQMVK Sbjct: 245 DGALNVDLTEFQTNLVPYPRIHFPLATYAPVISAEKAYHEQLTVAEITNACFEPANQMVK 304 Query: 903 CDPRNGKYMSCCLLYRGDVVPKDVXXXXXXXXXXXXXQFVDWCPTGFKVGINYQPPTVIP 1082 CDPR+GKYMSCC+LYRGDVVPKDV QFVDWCPTGFKVGINYQPPTV+P Sbjct: 305 CDPRHGKYMSCCMLYRGDVVPKDVNAAIATIKTKRTIQFVDWCPTGFKVGINYQPPTVVP 364 Query: 1083 GGDMAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYSKRAFVHWYVGEGMEEGEFSEARED 1262 GGD+AKVQRAVCMLSNTTAIAEAWARLDHKFDLMY+KRAFVHWYVGEGMEEGEFSEARED Sbjct: 365 GGDLAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYAKRAFVHWYVGEGMEEGEFSEARED 424 Query: 1263 LAALEKDYEEVGM 1301 LAALEKDYEEVG+ Sbjct: 425 LAALEKDYEEVGV 437
>sp|P18258|TBA1_PARLI Tubulin alpha-1 chain Length = 452 Score = 759 bits (1961), Expect = 0.0 Identities = 365/433 (84%), Positives = 392/433 (90%) Frame = +3 Query: 3 ISIHIGQAGVQMGNACWELYCLEHGIQQDGQMPSEKPKHGNEDCFNTFFSETGNGKYVPR 182 ISIH+GQAGVQMGNACWELYCLEHGIQ DGQMPS+K G +D FNTFFSETG GK+VPR Sbjct: 5 ISIHVGQAGVQMGNACWELYCLEHGIQPDGQMPSDKTIGGGDDSFNTFFSETGAGKHVPR 64 Query: 183 ALFVDLEPSVIDEVRNGAYRQLFHPEQLISGKEDAANNYARGHYTVGKELIDQVLDRIRK 362 A+FVDLEP+V+DEVR G YRQLFHPEQLI+GKEDAANNYARGHYTVGKELID VLDRIRK Sbjct: 65 AVFVDLEPTVVDEVRTGTYRQLFHPEQLITGKEDAANNYARGHYTVGKELIDIVLDRIRK 124 Query: 363 VADNCTGLQGFLMFHXXXXXXXXXXXXLLMERLSVDYGKKSKLEFAVYPAPQIATAVVEP 542 +AD CTGLQGFL+FH LLMERLSVDYGKKSKLEFAVYPAPQI+TAVVEP Sbjct: 125 LADQCTGLQGFLIFHSFGGGTGSGFSSLLMERLSVDYGKKSKLEFAVYPAPQISTAVVEP 184 Query: 543 YNSILTTHTTLEHSDCAFMVDNEAIYDICKKNLGIERPTYTSLNRLIGQIVSSITASLRF 722 YNSILTTHTTLEHSDCAFMVDNEAIYDIC++NL IERPTYT+LNRLIGQIVSSITASLRF Sbjct: 185 YNSILTTHTTLEHSDCAFMVDNEAIYDICRRNLDIERPTYTNLNRLIGQIVSSITASLRF 244 Query: 723 DGALNVDLNEFQTNLVPYPRIHFPLATYAPIICADRAFHEQLSVAEITNSCFEHNNQMVK 902 DGALNVDL EFQTNLVPYPRIHFPLATYAP+I A++A+HEQL+V+EITN+CFE NQMVK Sbjct: 245 DGALNVDLTEFQTNLVPYPRIHFPLATYAPVISAEKAYHEQLTVSEITNACFEPANQMVK 304 Query: 903 CDPRNGKYMSCCLLYRGDVVPKDVXXXXXXXXXXXXXQFVDWCPTGFKVGINYQPPTVIP 1082 CDPR+GKYM+CCLLYRGDVVPKDV QFVDWCPTGFKVGINYQPPTV+P Sbjct: 305 CDPRHGKYMACCLLYRGDVVPKDVNAAIATIKTKRTIQFVDWCPTGFKVGINYQPPTVVP 364 Query: 1083 GGDMAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYSKRAFVHWYVGEGMEEGEFSEARED 1262 GGD+AKVQRAVCMLSNTTAIAEAWARLDHKFDLMY+KRAFVHWYVGEGMEEGEFSEARED Sbjct: 365 GGDLAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYAKRAFVHWYVGEGMEEGEFSEARED 424 Query: 1263 LAALEKDYEEVGM 1301 LAALEKDYEEVG+ Sbjct: 425 LAALEKDYEEVGV 437
>sp|Q91060|TBA_NOTVI Tubulin alpha chain Length = 450 Score = 759 bits (1959), Expect = 0.0 Identities = 361/433 (83%), Positives = 391/433 (90%) Frame = +3 Query: 3 ISIHIGQAGVQMGNACWELYCLEHGIQQDGQMPSEKPKHGNEDCFNTFFSETGNGKYVPR 182 IS+H+GQAGVQMGNACWELYCLEHGIQ DGQMPS+K G +D FNTFFSETG GK+VPR Sbjct: 5 ISVHVGQAGVQMGNACWELYCLEHGIQPDGQMPSDKTIGGGDDSFNTFFSETGAGKHVPR 64 Query: 183 ALFVDLEPSVIDEVRNGAYRQLFHPEQLISGKEDAANNYARGHYTVGKELIDQVLDRIRK 362 A+FVDLEPSVIDEVR G YRQLFHPEQLISGKEDAANNYARGHYT+GKE+IDQVLDR+RK Sbjct: 65 AIFVDLEPSVIDEVRTGTYRQLFHPEQLISGKEDAANNYARGHYTIGKEIIDQVLDRMRK 124 Query: 363 VADNCTGLQGFLMFHXXXXXXXXXXXXLLMERLSVDYGKKSKLEFAVYPAPQIATAVVEP 542 +AD CTGLQGFL+FH LLMERLSVDYGKKSKLEFA+YPAPQ++TAVVEP Sbjct: 125 LADQCTGLQGFLVFHSFGGGTGSGFTSLLMERLSVDYGKKSKLEFAIYPAPQVSTAVVEP 184 Query: 543 YNSILTTHTTLEHSDCAFMVDNEAIYDICKKNLGIERPTYTSLNRLIGQIVSSITASLRF 722 YNSILTTHTTLEHSDCAFMVDNEAIYDIC++NL IERP+YT+LNRLI QIVSSITASLRF Sbjct: 185 YNSILTTHTTLEHSDCAFMVDNEAIYDICRRNLDIERPSYTNLNRLISQIVSSITASLRF 244 Query: 723 DGALNVDLNEFQTNLVPYPRIHFPLATYAPIICADRAFHEQLSVAEITNSCFEHNNQMVK 902 DGALNVDL EFQTNLVPYPRIHFPLATYAP+I A++A+HEQLSVAEITN+CFE NQMVK Sbjct: 245 DGALNVDLTEFQTNLVPYPRIHFPLATYAPVISAEKAYHEQLSVAEITNACFEPANQMVK 304 Query: 903 CDPRNGKYMSCCLLYRGDVVPKDVXXXXXXXXXXXXXQFVDWCPTGFKVGINYQPPTVIP 1082 CDPR+GKYM+CCLLYRGDVVPKDV QFVDWCPTGFKVGINYQPPT +P Sbjct: 305 CDPRHGKYMACCLLYRGDVVPKDVNAAIAAIKTKRSIQFVDWCPTGFKVGINYQPPTAVP 364 Query: 1083 GGDMAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYSKRAFVHWYVGEGMEEGEFSEARED 1262 GGD+AKVQRAVCMLSNTTAIAEAWARLDHKFDLMY+KRAFVHWYVGEGMEEGEFSEARED Sbjct: 365 GGDLAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYAKRAFVHWYVGEGMEEGEFSEARED 424 Query: 1263 LAALEKDYEEVGM 1301 +AALEKDYEEVG+ Sbjct: 425 MAALEKDYEEVGL 437
>sp|P06603|TBA1_DROME Tubulin alpha-1 chain Length = 450 Score = 758 bits (1957), Expect = 0.0 Identities = 360/433 (83%), Positives = 391/433 (90%) Frame = +3 Query: 3 ISIHIGQAGVQMGNACWELYCLEHGIQQDGQMPSEKPKHGNEDCFNTFFSETGNGKYVPR 182 ISIH+GQAGVQ+GNACWELYCLEHGIQ DGQMPS+K G +D FNTFFSETG GK+VPR Sbjct: 5 ISIHVGQAGVQIGNACWELYCLEHGIQPDGQMPSDKTVGGGDDSFNTFFSETGAGKHVPR 64 Query: 183 ALFVDLEPSVIDEVRNGAYRQLFHPEQLISGKEDAANNYARGHYTVGKELIDQVLDRIRK 362 A+FVDLEP+V+DEVR G YRQLFHPEQLI+GKEDAANNYARGHYT+GKE++D VLDRIRK Sbjct: 65 AVFVDLEPTVVDEVRTGTYRQLFHPEQLITGKEDAANNYARGHYTIGKEIVDLVLDRIRK 124 Query: 363 VADNCTGLQGFLMFHXXXXXXXXXXXXLLMERLSVDYGKKSKLEFAVYPAPQIATAVVEP 542 +AD CTGLQGFL+FH LLMERLSVDYGKKSKLEFA+YPAPQ++TAVVEP Sbjct: 125 LADQCTGLQGFLIFHSFGGGTGSGFTSLLMERLSVDYGKKSKLEFAIYPAPQVSTAVVEP 184 Query: 543 YNSILTTHTTLEHSDCAFMVDNEAIYDICKKNLGIERPTYTSLNRLIGQIVSSITASLRF 722 YNSILTTHTTLEHSDCAFMVDNEAIYDIC++NL IERPTYT+LNRLIGQIVSSITASLRF Sbjct: 185 YNSILTTHTTLEHSDCAFMVDNEAIYDICRRNLDIERPTYTNLNRLIGQIVSSITASLRF 244 Query: 723 DGALNVDLNEFQTNLVPYPRIHFPLATYAPIICADRAFHEQLSVAEITNSCFEHNNQMVK 902 DGALNVDL EFQTNLVPYPRIHFPL TYAP+I A++A+HEQLSVAEITN+CFE NQMVK Sbjct: 245 DGALNVDLTEFQTNLVPYPRIHFPLVTYAPVISAEKAYHEQLSVAEITNACFEPANQMVK 304 Query: 903 CDPRNGKYMSCCLLYRGDVVPKDVXXXXXXXXXXXXXQFVDWCPTGFKVGINYQPPTVIP 1082 CDPR+GKYM+CC+LYRGDVVPKDV QFVDWCPTGFKVGINYQPPTV+P Sbjct: 305 CDPRHGKYMACCMLYRGDVVPKDVNAAIATIKTKRTIQFVDWCPTGFKVGINYQPPTVVP 364 Query: 1083 GGDMAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYSKRAFVHWYVGEGMEEGEFSEARED 1262 GGD+AKVQRAVCMLSNTTAIAEAWARLDHKFDLMY+KRAFVHWYVGEGMEEGEFSEARED Sbjct: 365 GGDLAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYAKRAFVHWYVGEGMEEGEFSEARED 424 Query: 1263 LAALEKDYEEVGM 1301 LAALEKDYEEVGM Sbjct: 425 LAALEKDYEEVGM 437
>sp|P68369|TBA1_MOUSE Tubulin alpha-1 chain (Alpha-tubulin 1) (Alpha-tubulin isotype M-alpha-1) sp|P68362|TBA2_CRIGR Tubulin alpha-2 chain (Alpha-tubulin 2) (Alpha-tubulin II) sp|Q71U36|TBA3_HUMAN Tubulin alpha-3 chain (Alpha-tubulin 3) (Tubulin B-alpha-1) sp|P68370|TBA1_RAT Tubulin alpha-1 chain (Alpha-tubulin 1) Length = 451 Score = 757 bits (1955), Expect = 0.0 Identities = 361/433 (83%), Positives = 392/433 (90%) Frame = +3 Query: 3 ISIHIGQAGVQMGNACWELYCLEHGIQQDGQMPSEKPKHGNEDCFNTFFSETGNGKYVPR 182 ISIH+GQAGVQ+GNACWELYCLEHGIQ DGQMPS+K G +D FNTFFSETG GK+VPR Sbjct: 5 ISIHVGQAGVQIGNACWELYCLEHGIQPDGQMPSDKTIGGGDDSFNTFFSETGAGKHVPR 64 Query: 183 ALFVDLEPSVIDEVRNGAYRQLFHPEQLISGKEDAANNYARGHYTVGKELIDQVLDRIRK 362 A+FVDLEP+VIDEVR G YRQLFHPEQLI+GKEDAANNYARGHYT+GKE+ID VLDRIRK Sbjct: 65 AVFVDLEPTVIDEVRTGTYRQLFHPEQLITGKEDAANNYARGHYTIGKEIIDLVLDRIRK 124 Query: 363 VADNCTGLQGFLMFHXXXXXXXXXXXXLLMERLSVDYGKKSKLEFAVYPAPQIATAVVEP 542 +AD CTGLQGFL+FH LLMERLSVDYGKKSKLEF++YPAPQ++TAVVEP Sbjct: 125 LADQCTGLQGFLVFHSFGGGTGSGFTSLLMERLSVDYGKKSKLEFSIYPAPQVSTAVVEP 184 Query: 543 YNSILTTHTTLEHSDCAFMVDNEAIYDICKKNLGIERPTYTSLNRLIGQIVSSITASLRF 722 YNSILTTHTTLEHSDCAFMVDNEAIYDIC++NL IERPTYT+LNRLIGQIVSSITASLRF Sbjct: 185 YNSILTTHTTLEHSDCAFMVDNEAIYDICRRNLDIERPTYTNLNRLIGQIVSSITASLRF 244 Query: 723 DGALNVDLNEFQTNLVPYPRIHFPLATYAPIICADRAFHEQLSVAEITNSCFEHNNQMVK 902 DGALNVDL EFQTNLVPYPRIHFPLATYAP+I A++A+HEQLSVAEITN+CFE NQMVK Sbjct: 245 DGALNVDLTEFQTNLVPYPRIHFPLATYAPVISAEKAYHEQLSVAEITNACFEPANQMVK 304 Query: 903 CDPRNGKYMSCCLLYRGDVVPKDVXXXXXXXXXXXXXQFVDWCPTGFKVGINYQPPTVIP 1082 CDPR+GKYM+CCLLYRGDVVPKDV QFVDWCPTGFKVGINYQPPTV+P Sbjct: 305 CDPRHGKYMACCLLYRGDVVPKDVNAAIATIKTKRTIQFVDWCPTGFKVGINYQPPTVVP 364 Query: 1083 GGDMAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYSKRAFVHWYVGEGMEEGEFSEARED 1262 GGD+AKVQRAVCMLSNTTAIAEAWARLDHKFDLMY+KRAFVHWYVGEGMEEGEFSEARED Sbjct: 365 GGDLAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYAKRAFVHWYVGEGMEEGEFSEARED 424 Query: 1263 LAALEKDYEEVGM 1301 +AALEKDYEEVG+ Sbjct: 425 MAALEKDYEEVGV 437
>sp|Q8T6A5|TBA1_APLCA Tubulin alpha-1 chain Length = 451 Score = 757 bits (1955), Expect = 0.0 Identities = 362/433 (83%), Positives = 393/433 (90%) Frame = +3 Query: 3 ISIHIGQAGVQMGNACWELYCLEHGIQQDGQMPSEKPKHGNEDCFNTFFSETGNGKYVPR 182 ISIH+GQAGVQ+GNACWELYCLEHGIQ DGQMPS+K G +D FNTFFSETG GK+VPR Sbjct: 5 ISIHVGQAGVQIGNACWELYCLEHGIQPDGQMPSDKTIGGGDDSFNTFFSETGAGKHVPR 64 Query: 183 ALFVDLEPSVIDEVRNGAYRQLFHPEQLISGKEDAANNYARGHYTVGKELIDQVLDRIRK 362 A+FVDLEPSV+DEVR+G YRQLFHPEQLI+GKEDAANNYARGHYTVGKELID VLDRIRK Sbjct: 65 AVFVDLEPSVVDEVRSGTYRQLFHPEQLITGKEDAANNYARGHYTVGKELIDLVLDRIRK 124 Query: 363 VADNCTGLQGFLMFHXXXXXXXXXXXXLLMERLSVDYGKKSKLEFAVYPAPQIATAVVEP 542 +AD CTGLQGFL+FH LLMERLSVDYGKKSKLEF+VYPAPQ++TAVVEP Sbjct: 125 LADQCTGLQGFLIFHSFGGGTGSGFTSLLMERLSVDYGKKSKLEFSVYPAPQVSTAVVEP 184 Query: 543 YNSILTTHTTLEHSDCAFMVDNEAIYDICKKNLGIERPTYTSLNRLIGQIVSSITASLRF 722 YNSILTTHTTLEHSDCAFMVDNEAIYDIC++NL IERP+YT+LNRLIGQIVSSITASLRF Sbjct: 185 YNSILTTHTTLEHSDCAFMVDNEAIYDICRRNLDIERPSYTNLNRLIGQIVSSITASLRF 244 Query: 723 DGALNVDLNEFQTNLVPYPRIHFPLATYAPIICADRAFHEQLSVAEITNSCFEHNNQMVK 902 DGALNVDL EFQTNLVPYPRIHFPLATYAP+I A++A+HEQLSVAEITN+CFE NQ+VK Sbjct: 245 DGALNVDLTEFQTNLVPYPRIHFPLATYAPVISAEKAYHEQLSVAEITNACFEPANQLVK 304 Query: 903 CDPRNGKYMSCCLLYRGDVVPKDVXXXXXXXXXXXXXQFVDWCPTGFKVGINYQPPTVIP 1082 CDPR+GKYM+CC+LYRGDVVPKDV QFVDWCPTGFKVGINYQPPTV+P Sbjct: 305 CDPRHGKYMACCMLYRGDVVPKDVNAAIATIKTKRTIQFVDWCPTGFKVGINYQPPTVVP 364 Query: 1083 GGDMAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYSKRAFVHWYVGEGMEEGEFSEARED 1262 GGD+AKVQRAVCMLSNTTAIAEAWARLDHKFDLMY+KRAFVHWYVGEGMEEGEFSEARED Sbjct: 365 GGDLAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYAKRAFVHWYVGEGMEEGEFSEARED 424 Query: 1263 LAALEKDYEEVGM 1301 LAALEKDYEEVG+ Sbjct: 425 LAALEKDYEEVGV 437
>sp|P05214|TBA3_MOUSE Tubulin alpha-3/alpha-7 chain (Alpha-tubulin 3/7) (Alpha-tubulin isotype M-alpha-3/7) sp|Q68FR8|TBA3_RAT Tubulin alpha-3 chain (Alpha-tubulin 3) sp|Q13748|TBA2_HUMAN Tubulin alpha-2 chain (Alpha-tubulin 2) Length = 450 Score = 756 bits (1953), Expect = 0.0 Identities = 360/433 (83%), Positives = 392/433 (90%) Frame = +3 Query: 3 ISIHIGQAGVQMGNACWELYCLEHGIQQDGQMPSEKPKHGNEDCFNTFFSETGNGKYVPR 182 ISIH+GQAGVQ+GNACWELYCLEHGIQ DGQMPS+K G +D FNTFFSETG GK+VPR Sbjct: 5 ISIHVGQAGVQIGNACWELYCLEHGIQPDGQMPSDKTIGGGDDSFNTFFSETGAGKHVPR 64 Query: 183 ALFVDLEPSVIDEVRNGAYRQLFHPEQLISGKEDAANNYARGHYTVGKELIDQVLDRIRK 362 A+FVDLEP+V+DEVR G YRQLFHPEQLI+GKEDAANNYARGHYT+GKE++D VLDRIRK Sbjct: 65 AVFVDLEPTVVDEVRTGTYRQLFHPEQLITGKEDAANNYARGHYTIGKEIVDLVLDRIRK 124 Query: 363 VADNCTGLQGFLMFHXXXXXXXXXXXXLLMERLSVDYGKKSKLEFAVYPAPQIATAVVEP 542 +AD CTGLQGFL+FH LLMERLSVDYGKKSKLEFA+YPAPQ++TAVVEP Sbjct: 125 LADLCTGLQGFLIFHSFGGGTGSGFASLLMERLSVDYGKKSKLEFAIYPAPQVSTAVVEP 184 Query: 543 YNSILTTHTTLEHSDCAFMVDNEAIYDICKKNLGIERPTYTSLNRLIGQIVSSITASLRF 722 YNSILTTHTTLEHSDCAFMVDNEAIYDIC++NL IERPTYT+LNRLIGQIVSSITASLRF Sbjct: 185 YNSILTTHTTLEHSDCAFMVDNEAIYDICRRNLDIERPTYTNLNRLIGQIVSSITASLRF 244 Query: 723 DGALNVDLNEFQTNLVPYPRIHFPLATYAPIICADRAFHEQLSVAEITNSCFEHNNQMVK 902 DGALNVDL EFQTNLVPYPRIHFPLATYAP+I A++A+HEQLSVAEITN+CFE NQMVK Sbjct: 245 DGALNVDLTEFQTNLVPYPRIHFPLATYAPVISAEKAYHEQLSVAEITNACFEPANQMVK 304 Query: 903 CDPRNGKYMSCCLLYRGDVVPKDVXXXXXXXXXXXXXQFVDWCPTGFKVGINYQPPTVIP 1082 CDPR+GKYM+CC+LYRGDVVPKDV QFVDWCPTGFKVGINYQPPTV+P Sbjct: 305 CDPRHGKYMACCMLYRGDVVPKDVNAAIATIKTKRTIQFVDWCPTGFKVGINYQPPTVVP 364 Query: 1083 GGDMAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYSKRAFVHWYVGEGMEEGEFSEARED 1262 GGD+AKVQRAVCMLSNTTAIAEAWARLDHKFDLMY+KRAFVHWYVGEGMEEGEFSEARED Sbjct: 365 GGDLAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYAKRAFVHWYVGEGMEEGEFSEARED 424 Query: 1263 LAALEKDYEEVGM 1301 LAALEKDYEEVG+ Sbjct: 425 LAALEKDYEEVGV 437
>sp|P68361|TBA1_CRIGR Tubulin alpha-1 chain (Alpha-tubulin 1) (Alpha-tubulin I) sp|P68363|TBAK_HUMAN Tubulin alpha-ubiquitous chain (Alpha-tubulin ubiquitous) (Tubulin K-alpha-1) sp|P05213|TBA2_MOUSE Tubulin alpha-2 chain (Alpha-tubulin 2) (Alpha-tubulin isotype M-alpha-2) sp|P68360|TBA1_MERUN Tubulin alpha-1 chain (Alpha-tubulin 1) sp|Q6P9V9|TBA2_RAT Tubulin alpha-2 chain (Alpha-tubulin 2) Length = 451 Score = 755 bits (1949), Expect = 0.0 Identities = 360/433 (83%), Positives = 391/433 (90%) Frame = +3 Query: 3 ISIHIGQAGVQMGNACWELYCLEHGIQQDGQMPSEKPKHGNEDCFNTFFSETGNGKYVPR 182 ISIH+GQAGVQ+GNACWELYCLEHGIQ DGQMPS+K G +D FNTFFSETG GK+VPR Sbjct: 5 ISIHVGQAGVQIGNACWELYCLEHGIQPDGQMPSDKTIGGGDDSFNTFFSETGAGKHVPR 64 Query: 183 ALFVDLEPSVIDEVRNGAYRQLFHPEQLISGKEDAANNYARGHYTVGKELIDQVLDRIRK 362 A+FVDLEP+VIDEVR G YRQLFHPEQLI+GKEDAANNYARGHYT+GKE+ID VLDRIRK Sbjct: 65 AVFVDLEPTVIDEVRTGTYRQLFHPEQLITGKEDAANNYARGHYTIGKEIIDLVLDRIRK 124 Query: 363 VADNCTGLQGFLMFHXXXXXXXXXXXXLLMERLSVDYGKKSKLEFAVYPAPQIATAVVEP 542 +AD CTGLQGFL+FH LLMERLSVDYGKKSKLEF++YPAPQ++TAVVEP Sbjct: 125 LADQCTGLQGFLVFHSFGGGTGSGFTSLLMERLSVDYGKKSKLEFSIYPAPQVSTAVVEP 184 Query: 543 YNSILTTHTTLEHSDCAFMVDNEAIYDICKKNLGIERPTYTSLNRLIGQIVSSITASLRF 722 YNSILTTHTTLEHSDCAFMVDNEAIYDIC++NL IERPTYT+LNRLI QIVSSITASLRF Sbjct: 185 YNSILTTHTTLEHSDCAFMVDNEAIYDICRRNLDIERPTYTNLNRLISQIVSSITASLRF 244 Query: 723 DGALNVDLNEFQTNLVPYPRIHFPLATYAPIICADRAFHEQLSVAEITNSCFEHNNQMVK 902 DGALNVDL EFQTNLVPYPRIHFPLATYAP+I A++A+HEQLSVAEITN+CFE NQMVK Sbjct: 245 DGALNVDLTEFQTNLVPYPRIHFPLATYAPVISAEKAYHEQLSVAEITNACFEPANQMVK 304 Query: 903 CDPRNGKYMSCCLLYRGDVVPKDVXXXXXXXXXXXXXQFVDWCPTGFKVGINYQPPTVIP 1082 CDPR+GKYM+CCLLYRGDVVPKDV QFVDWCPTGFKVGINYQPPTV+P Sbjct: 305 CDPRHGKYMACCLLYRGDVVPKDVNAAIATIKTKRSIQFVDWCPTGFKVGINYQPPTVVP 364 Query: 1083 GGDMAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYSKRAFVHWYVGEGMEEGEFSEARED 1262 GGD+AKVQRAVCMLSNTTAIAEAWARLDHKFDLMY+KRAFVHWYVGEGMEEGEFSEARED Sbjct: 365 GGDLAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYAKRAFVHWYVGEGMEEGEFSEARED 424 Query: 1263 LAALEKDYEEVGM 1301 +AALEKDYEEVG+ Sbjct: 425 MAALEKDYEEVGV 437
>sp|P06605|TBA3_DROME Tubulin alpha-3 chain Length = 450 Score = 754 bits (1948), Expect = 0.0 Identities = 360/433 (83%), Positives = 390/433 (90%) Frame = +3 Query: 3 ISIHIGQAGVQMGNACWELYCLEHGIQQDGQMPSEKPKHGNEDCFNTFFSETGNGKYVPR 182 ISIH+GQAGVQ+GNACWELYCLEHGIQ DGQMPS+K G +D FNTFFSETG GK+VPR Sbjct: 5 ISIHVGQAGVQIGNACWELYCLEHGIQPDGQMPSDKTVGGGDDSFNTFFSETGAGKHVPR 64 Query: 183 ALFVDLEPSVIDEVRNGAYRQLFHPEQLISGKEDAANNYARGHYTVGKELIDQVLDRIRK 362 A+FVDLEP+V+DEVR G YRQLFHPEQLI+GKEDAANNYARGHYT+GKE++D VLDRIRK Sbjct: 65 AVFVDLEPTVVDEVRTGTYRQLFHPEQLITGKEDAANNYARGHYTIGKEIVDLVLDRIRK 124 Query: 363 VADNCTGLQGFLMFHXXXXXXXXXXXXLLMERLSVDYGKKSKLEFAVYPAPQIATAVVEP 542 +AD CTGLQGFL+FH LLMERLSVDYGKKSKLEFAVYPAPQ++TAVVEP Sbjct: 125 LADQCTGLQGFLIFHSFGGGTGSGFTSLLMERLSVDYGKKSKLEFAVYPAPQVSTAVVEP 184 Query: 543 YNSILTTHTTLEHSDCAFMVDNEAIYDICKKNLGIERPTYTSLNRLIGQIVSSITASLRF 722 YNSILTTHTTLEHSDCAFMVDNEAIYDIC++NL IERPTYT+LNRLIGQIVSSITASLRF Sbjct: 185 YNSILTTHTTLEHSDCAFMVDNEAIYDICRRNLDIERPTYTNLNRLIGQIVSSITASLRF 244 Query: 723 DGALNVDLNEFQTNLVPYPRIHFPLATYAPIICADRAFHEQLSVAEITNSCFEHNNQMVK 902 DGALNVDL EFQTNLVPYPRIHFPL TYAP+I A++A+HEQLSVAEITN+CFE NQMVK Sbjct: 245 DGALNVDLTEFQTNLVPYPRIHFPLVTYAPVISAEKAYHEQLSVAEITNACFEPANQMVK 304 Query: 903 CDPRNGKYMSCCLLYRGDVVPKDVXXXXXXXXXXXXXQFVDWCPTGFKVGINYQPPTVIP 1082 DPR+GKYM+CC+LYRGDVVPKDV QFVDWCPTGFKVGINYQPPTV+P Sbjct: 305 VDPRHGKYMACCMLYRGDVVPKDVNAAIATIKTKRTIQFVDWCPTGFKVGINYQPPTVVP 364 Query: 1083 GGDMAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYSKRAFVHWYVGEGMEEGEFSEARED 1262 GGD+AKVQRAVCMLSNTTAIAEAWARLDHKFDLMY+KRAFVHWYVGEGMEEGEFSEARED Sbjct: 365 GGDLAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYAKRAFVHWYVGEGMEEGEFSEARED 424 Query: 1263 LAALEKDYEEVGM 1301 LAALEKDYEEVGM Sbjct: 425 LAALEKDYEEVGM 437
>sp|P52273|TBA_BOMMO Tubulin alpha chain Length = 450 Score = 754 bits (1948), Expect = 0.0 Identities = 358/433 (82%), Positives = 390/433 (90%) Frame = +3 Query: 3 ISIHIGQAGVQMGNACWELYCLEHGIQQDGQMPSEKPKHGNEDCFNTFFSETGNGKYVPR 182 IS+H+GQAGVQ+GNACWELYCLEHGIQ DGQMP++K G +D FNTFFSETG GK+VPR Sbjct: 5 ISVHVGQAGVQIGNACWELYCLEHGIQPDGQMPTDKTIGGGDDSFNTFFSETGAGKHVPR 64 Query: 183 ALFVDLEPSVIDEVRNGAYRQLFHPEQLISGKEDAANNYARGHYTVGKELIDQVLDRIRK 362 ALFVDLEP+V+DEVR G YRQLFHPEQLI+GKEDAANNYARGHYT+GKE++D VLDRIRK Sbjct: 65 ALFVDLEPTVVDEVRTGTYRQLFHPEQLITGKEDAANNYARGHYTIGKEIVDLVLDRIRK 124 Query: 363 VADNCTGLQGFLMFHXXXXXXXXXXXXLLMERLSVDYGKKSKLEFAVYPAPQIATAVVEP 542 +AD CTGLQGFL+FH LLMERLSVDYGKKSKLEFA+YPAPQ++TAVVEP Sbjct: 125 LADQCTGLQGFLIFHSFGGGTGSGFTSLLMERLSVDYGKKSKLEFAIYPAPQVSTAVVEP 184 Query: 543 YNSILTTHTTLEHSDCAFMVDNEAIYDICKKNLGIERPTYTSLNRLIGQIVSSITASLRF 722 YNSILTTHTTLEHSDCAFMVDNEAIYDIC++NL IE PTYT+LNRLIGQIVSSITASLRF Sbjct: 185 YNSILTTHTTLEHSDCAFMVDNEAIYDICRRNLDIEPPTYTNLNRLIGQIVSSITASLRF 244 Query: 723 DGALNVDLNEFQTNLVPYPRIHFPLATYAPIICADRAFHEQLSVAEITNSCFEHNNQMVK 902 DGALNVDL EFQTNLVPYPRIHFPL TYAP+I A++A+HEQLSVAEITN+CFE NQMVK Sbjct: 245 DGALNVDLTEFQTNLVPYPRIHFPLVTYAPVISAEKAYHEQLSVAEITNACFEPANQMVK 304 Query: 903 CDPRNGKYMSCCLLYRGDVVPKDVXXXXXXXXXXXXXQFVDWCPTGFKVGINYQPPTVIP 1082 CDPR+GKYM+CC+LYRGDVVPKDV QFVDWCPTGFKVGINYQPPTV+P Sbjct: 305 CDPRHGKYMACCMLYRGDVVPKDVNAAIATIKTKRTIQFVDWCPTGFKVGINYQPPTVVP 364 Query: 1083 GGDMAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYSKRAFVHWYVGEGMEEGEFSEARED 1262 GGD+AKVQRAVCMLSNTTAIAEAWARLDHKFDLMY+KRAFVHWYVGEGMEEGEFSEARED Sbjct: 365 GGDLAKVQRAVCMLSNTTAIAEAWARLDHKFDLMYAKRAFVHWYVGEGMEEGEFSEARED 424 Query: 1263 LAALEKDYEEVGM 1301 LAALEKDYEEVGM Sbjct: 425 LAALEKDYEEVGM 437
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 156,691,255
Number of Sequences: 369166
Number of extensions: 3243478
Number of successful extensions: 9389
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8408
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8888
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 16506021310
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail
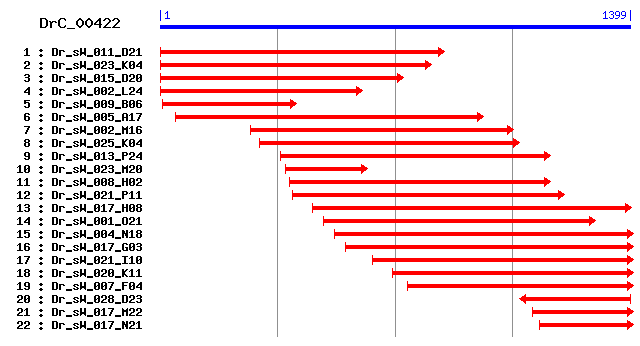
- Dr_sW_009_B06
- Dr_sW_002_L24
- Dr_sW_023_M20
- Dr_sW_015_D20
- Dr_sW_011_D21
- Dr_sW_023_K04
- Dr_sW_005_A17
- Dr_sW_002_M16
- Dr_sW_025_K04
- Dr_sW_013_P24
- Dr_sW_008_H02
- Dr_sW_021_P11
- Dr_sW_001_O21
- Dr_sW_017_H08
- Dr_sW_004_N18
- Dr_sW_007_F04
- Dr_sW_017_N21
- Dr_sW_017_M22
- Dr_sW_020_K11
- Dr_sW_028_D23
- Dr_sW_017_G03
- Dr_sW_021_I10