DrC_01050
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01050 (852 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P48060|GLIP1_HUMAN Glioma pathogenesis-related protein 1... 138 2e-32 sp|P60623|CRVP_TRIST Cysteine-rich secretory protein precur... 132 1e-30 sp|Q9CWG1|GLIP1_MOUSE Glioma pathogenesis-related protein 1... 129 1e-29 sp|O19010|CRIS3_HORSE Cysteine-rich secretory protein 3 pre... 127 4e-29 sp|Q8JI39|CRVP_TRIFL Triflin precursor 124 4e-28 sp|Q7ZZN9|CRVP_TRIJE Cysteine-rich venom protein precursor ... 123 7e-28 sp|Q8JGT9|CRVP_RHATT Tigrin precursor 122 2e-27 sp|Q8JI40|CRVP_AGKHA Ablomin precursor 122 2e-27 sp|Q8AVA4|CRVP_PSEAU Pseudechetoxin precursor (PsTx) 121 2e-27 sp|P79845|CRVP_TRIMU Cysteine-rich venom protein precursor ... 120 6e-27
>sp|P48060|GLIP1_HUMAN Glioma pathogenesis-related protein 1 precursor (GliPR 1) (RTVP-1 protein) Length = 266 Score = 138 bits (348), Expect = 2e-32 Identities = 80/204 (39%), Positives = 113/204 (55%), Gaps = 14/204 (6%) Frame = +2 Query: 17 IVSFIS-IALTFSTPP--EYKDFL---LKAHNDLRRDVALGKTPNQPGAANMIEMVWDDA 178 +VSF+S + T + P E +DF+ ++ HN R +V +P A++M+ M WD A Sbjct: 11 MVSFVSNYSHTANILPDIENEDFIKDCVRIHNKFRSEV-------KPTASDMLYMTWDPA 63 Query: 179 LAGKAEEWAGKCIAGHDTY---DDRQLDKFMFVGQNMFAGS----NYKEVVQGWYDEYKD 337 LA A+ WA C H+T + F +G+N++ GS + + WYDE +D Sbjct: 64 LAQIAKAWASNCQFSHNTRLKPPHKLHPNFTSLGENIWTGSVPIFSVSSAITNWYDEIQD 123 Query: 338 YTYEGKGCSAVCGHYTQVAWAKSYAVGCAAVNCKEKTG-GSFAYGWLFICNYGPAGNFND 514 Y ++ + C VCGHYTQV WA SY VGCA C + +G + + G FICNYGP GN+ Sbjct: 124 YDFKTRICKKVCGHYTQVVWADSYKVGCAVQFCPKVSGFDALSNGAHFICNYGPGGNY-P 182 Query: 515 EAPYEVGAACSKCPKGTSCKNNLC 586 PY+ GA CS CP C +NLC Sbjct: 183 TWPYKRGATCSACPNNDKCLDNLC 206
>sp|P60623|CRVP_TRIST Cysteine-rich secretory protein precursor (Stecrisp) Length = 233 Score = 132 bits (332), Expect = 1e-30 Identities = 71/184 (38%), Positives = 97/184 (52%), Gaps = 8/184 (4%) Frame = +2 Query: 59 PEYKDFLLKAHNDLRRDVALGKTPNQPGAANMIEMVWDDALAGKAEEWAGKCIAGHDTYD 238 PE ++ ++ HN LRR V P A+NM+ M W A AE WA +CI H +Y+ Sbjct: 24 PEIQNEIVDLHNSLRRSV-------NPTASNMLRMEWYPEAADNAERWAYRCIESHSSYE 76 Query: 239 DRQLDKFMFVGQNMFAGS---NYKEVVQGWYDEYKDYTYEGKGC---SAVCGHYTQVAWA 400 R ++ G+N++ + +++ W+DEYKD+ Y G G +AV GHYTQ+ W Sbjct: 77 SRVIEGIK-CGENIYMSPYPMKWTDIIHAWHDEYKDFKY-GVGADPPNAVTGHYTQIVWY 134 Query: 401 KSYAVGCAAVNCKEKTGGSFAYGWLFICNYGPAGNF--NDEAPYEVGAACSKCPKGTSCK 574 KSY +GCAA C S Y + F+C Y PAGNF PY G C CP + C Sbjct: 135 KSYRIGCAAAYCP-----SSPYSYFFVCQYCPAGNFIGKTATPYTSGTPCGDCP--SDCD 187 Query: 575 NNLC 586 N LC Sbjct: 188 NGLC 191
>sp|Q9CWG1|GLIP1_MOUSE Glioma pathogenesis-related protein 1 precursor (GliPR 1) Length = 255 Score = 129 bits (323), Expect = 1e-29 Identities = 77/198 (38%), Positives = 101/198 (51%), Gaps = 10/198 (5%) Frame = +2 Query: 23 SFISIALTFSTPPEY--KDFL---LKAHNDLRRDVALGKTPNQPGAANMIEMVWDDALAG 187 S S + T ST P+ +DF+ ++ HN LR V+ P A NM+ M WD LA Sbjct: 14 SVSSSSFTASTLPDITNEDFIKECVQVHNQLRSKVS-------PPARNMLYMSWDPKLAQ 66 Query: 188 KAEEWAGKCIAGHDTY-DDRQLDKFMFVGQNMFAGS----NYKEVVQGWYDEYKDYTYEG 352 A+ W C H+ R F +G+N++ GS + + WY+E K Y + Sbjct: 67 IAKAWTKSCEFKHNPQLHSRIHPNFTALGENIWLGSLSIFSVSSAISAWYEEIKHYDFST 126 Query: 353 KGCSAVCGHYTQVAWAKSYAVGCAAVNCKEKTGGSFAYGWLFICNYGPAGNFNDEAPYEV 532 + C VCGHYTQV WA SY +GCA C G FIC+YGPAGN+ PY+ Sbjct: 127 RKCRHVCGHYTQVVWADSYKLGCAVQLCPN--------GANFICDYGPAGNY-PTWPYKQ 177 Query: 533 GAACSKCPKGTSCKNNLC 586 GA CS CPK C N+LC Sbjct: 178 GATCSDCPKDDKCLNSLC 195
>sp|O19010|CRIS3_HORSE Cysteine-rich secretory protein 3 precursor (CRISP-3) Length = 245 Score = 127 bits (319), Expect = 4e-29 Identities = 71/185 (38%), Positives = 102/185 (55%), Gaps = 7/185 (3%) Frame = +2 Query: 53 TPPEYKDFLLKAHNDLRRDVALGKTPNQPGAANMIEMVWDDALAGKAEEWAGKCIAGHDT 232 T E + ++ HNDLRR V+ P A+NM++M WD A A+ WA KC+ H Sbjct: 33 TKSEVQKEIVNKHNDLRRTVS-------PLASNMLKMQWDSKTATNAQNWANKCLLQHSK 85 Query: 233 YDDRQLDKFMFVGQNMFAGS---NYKEVVQGWYDEYKDYTY--EGKGCSAVCGHYTQVAW 397 +DR + M G+N+F S ++ + +Q W+DE D+ Y K +AV GHYTQV W Sbjct: 86 AEDRAVGT-MKCGENLFMSSIPNSWSDAIQNWHDEVHDFKYGVGPKTPNAVVGHYTQVVW 144 Query: 398 AKSYAVGCAAVNCKEKTGGSFAYGWLFICNYGPAGNFNDE--APYEVGAACSKCPKGTSC 571 SY VGC C ++ G+ Y ++C Y PAGN+ ++ PYE G C++CP +C Sbjct: 145 YSSYRVGCGIAYCPKQ--GTLKY--YYVCQYCPAGNYVNKINTPYEQGTPCARCP--GNC 198 Query: 572 KNNLC 586 N LC Sbjct: 199 DNGLC 203
>sp|Q8JI39|CRVP_TRIFL Triflin precursor Length = 240 Score = 124 bits (310), Expect = 4e-28 Identities = 68/184 (36%), Positives = 94/184 (51%), Gaps = 8/184 (4%) Frame = +2 Query: 59 PEYKDFLLKAHNDLRRDVALGKTPNQPGAANMIEMVWDDALAGKAEEWAGKCIAGHDTYD 238 PE ++ ++ HN LRR V P A+NM++M W A AE WA +CI H + D Sbjct: 31 PEIQNEIIDLHNSLRRSV-------NPTASNMLKMEWYPEAAANAERWAYRCIESHSSRD 83 Query: 239 DRQLDKFMFVGQNMFAGS---NYKEVVQGWYDEYKDYTYEGKGC---SAVCGHYTQVAWA 400 R + G+N++ + + +++ W+ EYKD+ Y G G AV GHYTQ+ W Sbjct: 84 SRVIGGIK-CGENIYMATYPAKWTDIIHAWHGEYKDFKY-GVGAVPSDAVIGHYTQIVWY 141 Query: 401 KSYAVGCAAVNCKEKTGGSFAYGWLFICNYGPAGNF--NDEAPYEVGAACSKCPKGTSCK 574 KSY GCAA C S Y + ++C Y PAGN PY+ G C CP + C Sbjct: 142 KSYRAGCAAAYCP-----SSKYSYFYVCQYCPAGNIIGKTATPYKSGPPCGDCP--SDCD 194 Query: 575 NNLC 586 N LC Sbjct: 195 NGLC 198
>sp|Q7ZZN9|CRVP_TRIJE Cysteine-rich venom protein precursor (TJ-CRVP) Length = 240 Score = 123 bits (308), Expect = 7e-28 Identities = 68/184 (36%), Positives = 93/184 (50%), Gaps = 8/184 (4%) Frame = +2 Query: 59 PEYKDFLLKAHNDLRRDVALGKTPNQPGAANMIEMVWDDALAGKAEEWAGKCIAGHDTYD 238 PE ++ ++ HN LRR V P A+NM++M W A AE WA CI H + D Sbjct: 31 PEIQNEIIDLHNSLRRSV-------NPTASNMLKMEWYPEAAANAERWAYGCIESHSSRD 83 Query: 239 DRQLDKFMFVGQNMFAGS---NYKEVVQGWYDEYKDYTYEGKGC---SAVCGHYTQVAWA 400 R ++ G+N++ + +++ W+ EYKD+ Y G G AV GHYTQ+ W Sbjct: 84 SRVIEGIK-CGENIYMSPYPMKWTDIIHAWHGEYKDFKY-GVGAVPSDAVVGHYTQIVWY 141 Query: 401 KSYAVGCAAVNCKEKTGGSFAYGWLFICNYGPAGNF--NDEAPYEVGAACSKCPKGTSCK 574 KSY +GCAA C S Y + ++C Y PAGN PY G C CP + C Sbjct: 142 KSYRIGCAAAYCP-----SAEYSYFYVCQYCPAGNMIGKTATPYTSGPPCGDCP--SDCD 194 Query: 575 NNLC 586 N LC Sbjct: 195 NGLC 198
>sp|Q8JGT9|CRVP_RHATT Tigrin precursor Length = 237 Score = 122 bits (305), Expect = 2e-27 Identities = 71/198 (35%), Positives = 101/198 (51%), Gaps = 10/198 (5%) Frame = +2 Query: 23 SFISIALTFSTP--PEYKDFLLKAHNDLRRDVALGKTPNQPGAANMIEMVWDDALAGKAE 196 SF ++ +P P + ++ HN RR V +P A NM++M W A AE Sbjct: 16 SFGNVDFNSESPRNPGKQQEIVNIHNSFRRSV-------RPTARNMLKMEWYSEAASNAE 68 Query: 197 EWAGKCIAGHDTYDDRQLDKFMFVGQNMFAGSN---YKEVVQGWYDEYKDYTYEGKGCS- 364 WA +C H R L+ G+N+F S+ + ++Q WYDEY+++ Y G G + Sbjct: 69 RWAYQCAYDHSPRSSRILNGIQ-CGENIFMASDPWAWTSIIQDWYDEYRNFVY-GVGANP 126 Query: 365 --AVCGHYTQVAWAKSYAVGCAAVNCKEKTGGSFAYGWLFICNYGPAGNF--NDEAPYEV 532 +V GHYTQ+ W KSY VGCAA C S Y + ++C Y PAGN + PY Sbjct: 127 PDSVTGHYTQIVWYKSYLVGCAAAYCP-----SSLYNYFYVCQYCPAGNIQGSTSTPYAS 181 Query: 533 GAACSKCPKGTSCKNNLC 586 G C+ CP ++C N LC Sbjct: 182 GPTCADCP--SNCDNGLC 197
>sp|Q8JI40|CRVP_AGKHA Ablomin precursor Length = 240 Score = 122 bits (305), Expect = 2e-27 Identities = 68/184 (36%), Positives = 97/184 (52%), Gaps = 8/184 (4%) Frame = +2 Query: 59 PEYKDFLLKAHNDLRRDVALGKTPNQPGAANMIEMVWDDALAGKAEEWAGKCIAGHDTYD 238 PE ++ ++ HN LRR V P A+NM++M W A AE WA +CI H + D Sbjct: 31 PEIQNEIVDLHNSLRRSV-------NPTASNMLKMEWYPEAAANAERWAYRCIEDHSSPD 83 Query: 239 DRQLDKFMFVGQNMFAGS---NYKEVVQGWYDEYKDYTYEGKGC---SAVCGHYTQVAWA 400 R L+ G+N++ + +++ W+DEYK++ Y G G +AV GH+TQ+ W Sbjct: 84 SRVLEGIK-CGENIYMSPIPMKWTDIIHIWHDEYKNFKY-GIGADPPNAVSGHFTQIVWY 141 Query: 401 KSYAVGCAAVNCKEKTGGSFAYGWLFICNYGPAGNFNDE--APYEVGAACSKCPKGTSCK 574 KSY GCAA C S Y + ++C Y PAGN + PY G C CP ++C Sbjct: 142 KSYRAGCAAAYCP-----SSEYSYFYVCQYCPAGNMRGKTATPYTSGPPCGDCP--SACD 194 Query: 575 NNLC 586 N LC Sbjct: 195 NGLC 198
>sp|Q8AVA4|CRVP_PSEAU Pseudechetoxin precursor (PsTx) Length = 238 Score = 121 bits (304), Expect = 2e-27 Identities = 68/186 (36%), Positives = 94/186 (50%), Gaps = 7/186 (3%) Frame = +2 Query: 50 STPPEYKDFLLKAHNDLRRDVALGKTPNQPGAANMIEMVWDDALAGKAEEWAGKCIAGHD 229 S Y+ ++ HN LRR V +P A NM++M W+ A A+ WA +C H Sbjct: 28 SNKKNYQKEIVDKHNALRRSV-------KPTARNMLQMKWNSRAAQNAKRWANRCTFAHS 80 Query: 230 TYDDRQLDKFMFVGQNMFAGSN---YKEVVQGWYDEYKDYTYE--GKGCSAVCGHYTQVA 394 + R + K G+N+F S + VVQ WYDE K++ Y K +V GHYTQV Sbjct: 81 PPNKRTVGKLR-CGENIFMSSQPFPWSGVVQAWYDEIKNFVYGIGAKPPGSVIGHYTQVV 139 Query: 395 WAKSYAVGCAAVNCKEKTGGSFAYGWLFICNYGPAGNFNDE--APYEVGAACSKCPKGTS 568 W KSY +GCA+ C +L++C Y PAGN PY+ G C+ CP ++ Sbjct: 140 WYKSYLIGCASAKCSSSK-------YLYVCQYCPAGNIRGSIATPYKSGPPCADCP--SA 190 Query: 569 CKNNLC 586 C N LC Sbjct: 191 CVNKLC 196
>sp|P79845|CRVP_TRIMU Cysteine-rich venom protein precursor (TM-CRVP) Length = 240 Score = 120 bits (300), Expect = 6e-27 Identities = 67/184 (36%), Positives = 93/184 (50%), Gaps = 8/184 (4%) Frame = +2 Query: 59 PEYKDFLLKAHNDLRRDVALGKTPNQPGAANMIEMVWDDALAGKAEEWAGKCIAGHDTYD 238 PE ++ ++ HN LRR V P A+NM++M W A AE WA +CI H + D Sbjct: 31 PEIQNEIIDLHNSLRRSV-------NPTASNMLKMEWYPEAAANAERWAYRCIESHSSRD 83 Query: 239 DRQLDKFMFVGQNMFAG---SNYKEVVQGWYDEYKDYTYEGKGC---SAVCGHYTQVAWA 400 R + G+N++ + + +++ W+ EYKD+ Y G G +A GHYTQ+ W Sbjct: 84 SRVIGGIK-CGENIYMSPYPAKWTDIIHAWHGEYKDFKY-GVGAVPSNAATGHYTQIVWY 141 Query: 401 KSYAVGCAAVNCKEKTGGSFAYGWLFICNYGPAGNF--NDEAPYEVGAACSKCPKGTSCK 574 KSY GCAA C S Y + ++C Y PAGN PY G C CP + C Sbjct: 142 KSYRGGCAAAYCP-----SSKYRYFYVCQYCPAGNMIGKTATPYTSGPPCGDCP--SDCD 194 Query: 575 NNLC 586 N LC Sbjct: 195 NGLC 198
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 91,617,657
Number of Sequences: 369166
Number of extensions: 1894498
Number of successful extensions: 6079
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5702
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5871
length of database: 68,354,980
effective HSP length: 109
effective length of database: 48,218,865
effective search space used: 8390082510
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail
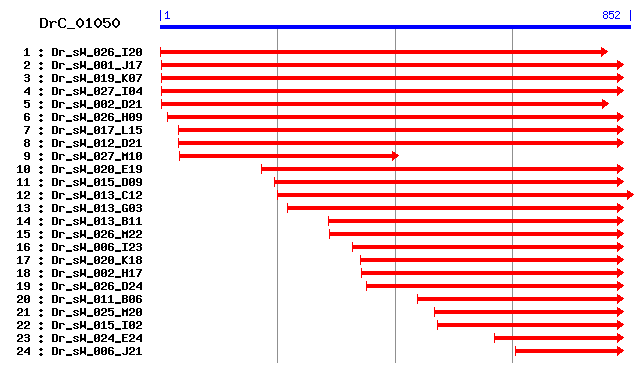
- Dr_sW_027_M10
- Dr_sW_026_I20
- Dr_sW_027_I04
- Dr_sW_001_J17
- Dr_sW_006_J21
- Dr_sW_024_E24
- Dr_sW_015_I02
- Dr_sW_025_M20
- Dr_sW_011_B06
- Dr_sW_026_D24
- Dr_sW_002_H17
- Dr_sW_020_K18
- Dr_sW_013_B11
- Dr_sW_026_M22
- Dr_sW_013_G03
- Dr_sW_015_D09
- Dr_sW_020_E19
- Dr_sW_012_D21
- Dr_sW_017_L15
- Dr_sW_002_D21
- Dr_sW_019_K07
- Dr_sW_026_H09
- Dr_sW_006_I23
- Dr_sW_013_C12