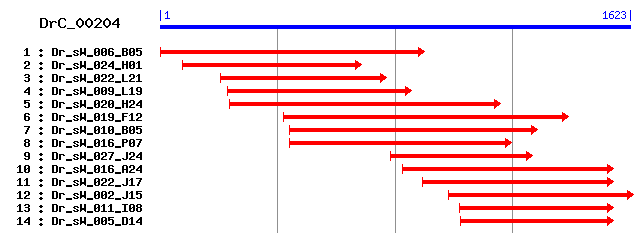
DrC_00204
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00204 (1619 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P00889|CISY_PIG Citrate synthase, mitochondrial precursor 659 0.0 sp|O75390|CISY_HUMAN Citrate synthase, mitochondrial precursor 653 0.0 sp|Q9CZU6|CISY_MOUSE Citrate synthase, mitochondrial precursor 652 0.0 sp|P23007|CISY_CHICK Citrate synthase, mitochondrial 626 e-179 sp|P34575|CISY_CAEEL Probable citrate synthase, mitochondri... 606 e-173 sp|P51044|CISY_ASPNG Citrate synthase, mitochondrial precursor 545 e-154 sp|O00098|CISY_EMENI Citrate synthase, mitochondrial precursor 544 e-154 sp|P00890|CISY1_YEAST Citrate synthase, mitochondrial precu... 537 e-152 sp|O80433|CISY_DAUCA Citrate synthase, mitochondrial precursor 536 e-152 sp|P20115|CISY4_ARATH Citrate synthase 4, mitochondrial pre... 536 e-152
>sp|P00889|CISY_PIG Citrate synthase, mitochondrial precursor Length = 464 Score = 659 bits (1700), Expect = 0.0 Identities = 313/434 (72%), Positives = 373/434 (85%) Frame = +3 Query: 111 ASTVNLKDILADTITKEQKKVSEFRKQHGETKVGEVTINMMYGGMRGIKGLVTETSVLDP 290 AS+ NLKDILAD I KEQ ++ FR+QHG T VG++T++MMYGGMRG+KGLV ETSVLDP Sbjct: 28 ASSTNLKDILADLIPKEQARIKTFRQQHGNTVVGQITVDMMYGGMRGMKGLVYETSVLDP 87 Query: 291 EEGIRFRGYSIPECQKVLPKSQGGEQPLPEGLFFLLLTGKIPTEEQVKWISKEWASRADL 470 +EGIRFRGYSIPECQK+LPK++GGE+PLPEGLF+LL+TG+IPTEEQV W+SKEWA RA L Sbjct: 88 DEGIRFRGYSIPECQKMLPKAKGGEEPLPEGLFWLLVTGQIPTEEQVSWLSKEWAKRAAL 147 Query: 471 PGHVVSMINNFPSSLHPMSQLVAASAALNSESKFVKAYHEGVSKSVYWETVYEDSMDLIA 650 P HVV+M++NFP++LHPMSQL AA ALNSES F +AY EG+ ++ YWE +YED MDLIA Sbjct: 148 PSHVVTMLDNFPTNLHPMSQLSAAITALNSESNFARAYAEGIHRTKYWELIYEDCMDLIA 207 Query: 651 KLSTVAAMIYRNLYRDGTSVGCIDNSKDWSENFVRMIGYEDPSFTELMRLYLTIHSDHEG 830 KL VAA IYRNLYR+G+S+G ID+ DWS NF M+GY D FTELMRLYLTIHSDHEG Sbjct: 208 KLPCVAAKIYRNLYREGSSIGAIDSKLDWSHNFTNMLGYTDAQFTELMRLYLTIHSDHEG 267 Query: 831 GNVSAHTCHLVGSALSDPYLSYAAAMCGLAGPLHGLANQEVLLWVQKMIEVIGTKPTDKD 1010 GNVSAHT HLVGSALSDPYLS+AAAM GLAGPLHGLANQEVL+W+ ++ + +G +D+ Sbjct: 268 GNVSAHTSHLVGSALSDPYLSFAAAMNGLAGPLHGLANQEVLVWLTQLQKEVGKDVSDEK 327 Query: 1011 IVEFIEKTLKAGQVVPGYGHAVLRKTDPRYMCQREFALKHLPDYPLFKVVSQLYQIVPDV 1190 + ++I TL +G+VVPGYGHAVLRKTDPRY CQREFALKHLP P+FK+V+QLY+IVP+V Sbjct: 328 LRDYIWNTLNSGRVVPGYGHAVLRKTDPRYTCQREFALKHLPHDPMFKLVAQLYKIVPNV 387 Query: 1191 LGKKSKIANPFPNVDAHSGVLLQYYGMTETNYYTVLFGISRALGILSSLIWDRALGLPIE 1370 L ++ K NP+PNVDAHSGVLLQYYGMTE NYYTVLFG+SRALG+L+ LIW RALG P+E Sbjct: 388 LLEQGKAKNPWPNVDAHSGVLLQYYGMTEMNYYTVLFGVSRALGVLAQLIWSRALGFPLE 447 Query: 1371 RPKSFSTDKLIELV 1412 RPKS STD LI+LV Sbjct: 448 RPKSMSTDGLIKLV 461
>sp|O75390|CISY_HUMAN Citrate synthase, mitochondrial precursor Length = 466 Score = 653 bits (1685), Expect = 0.0 Identities = 311/439 (70%), Positives = 377/439 (85%), Gaps = 1/439 (0%) Frame = +3 Query: 111 ASTVNLKDILADTITKEQKKVSEFRKQHGETKVGEVTINMMYGGMRGIKGLVTETSVLDP 290 AS+ NLKDILAD I KEQ ++ FR+QHG+T VG++T++MMYGGMRG+KGLV ETSVLDP Sbjct: 28 ASSTNLKDILADLIPKEQARIKTFRQQHGKTVVGQITVDMMYGGMRGMKGLVYETSVLDP 87 Query: 291 EEGIRFRGYSIPECQKVLPKSQGGEQPLPEGLFFLLLTGKIPTEEQVKWISKEWASRADL 470 +EGIRFRG+SIPECQK+LPK++GGE+PLPEGLF+LL+TG IPTEEQV W+SKEWA RA L Sbjct: 88 DEGIRFRGFSIPECQKLLPKAKGGEEPLPEGLFWLLVTGHIPTEEQVSWLSKEWAKRAAL 147 Query: 471 PGHVVSMINNFPSSLHPMSQLVAASAALNSESKFVKAYHEGVSKSVYWETVYEDSMDLIA 650 P HVV+M++NFP++LHPMSQL AA ALNSES F +AY +G+S++ YWE +YEDSMDLIA Sbjct: 148 PSHVVTMLDNFPTNLHPMSQLSAAVTALNSESNFARAYAQGISRTKYWELIYEDSMDLIA 207 Query: 651 KLSTVAAMIYRNLYRDGTSVGCIDNSKDWSENFVRMIGYEDPSFTELMRLYLTIHSDHEG 830 KL VAA IYRNLYR+G+ +G ID++ DWS NF M+GY D FTEL RLYLTIHSDHEG Sbjct: 208 KLPCVAAKIYRNLYREGSGIGAIDSNLDWSHNFTNMLGYTDHQFTELTRLYLTIHSDHEG 267 Query: 831 GNVSAHTCHLVGSALSDPYLSYAAAMCGLAGPLHGLANQEVLLWVQKMIEVIGTKPTDKD 1010 GNVSAHT HLVGSALSDPYLS+AAAM GLAGPLHGLANQEVL+W+ ++ + +G +D+ Sbjct: 268 GNVSAHTSHLVGSALSDPYLSFAAAMNGLAGPLHGLANQEVLVWLTQLQKEVGKDVSDEK 327 Query: 1011 IVEFIEKTLKAGQVVPGYGHAVLRKTDPRYMCQREFALKHLPDYPLFKVVSQLYQIVPDV 1190 + ++I TL +G+VVPGYGHAVLRKTDPRY CQREFALKHLP+ P+FK+V+QLY+IVP+V Sbjct: 328 LRDYIWNTLNSGRVVPGYGHAVLRKTDPRYTCQREFALKHLPNDPMFKLVAQLYKIVPNV 387 Query: 1191 LGKKSKIANPFPNVDAHSGVLLQYYGMTETNYYTVLFGISRALGILSSLIWDRALGLPIE 1370 L ++ K NP+PNVDAHSGVLLQYYGMTE NYYTVLFG+SRALG+L+ LIW RALG P+E Sbjct: 388 LLEQGKAKNPWPNVDAHSGVLLQYYGMTEMNYYTVLFGVSRALGVLAQLIWSRALGFPLE 447 Query: 1371 RPKSFSTDKLIELV-RKSG 1424 RPKS ST+ L++ V KSG Sbjct: 448 RPKSMSTEGLMKFVDSKSG 466
>sp|Q9CZU6|CISY_MOUSE Citrate synthase, mitochondrial precursor Length = 464 Score = 652 bits (1682), Expect = 0.0 Identities = 305/434 (70%), Positives = 375/434 (86%) Frame = +3 Query: 111 ASTVNLKDILADTITKEQKKVSEFRKQHGETKVGEVTINMMYGGMRGIKGLVTETSVLDP 290 AS+ NLKD+L++ I KEQ ++ F++QHG+T VG++T++MMYGGMRG+KGLV ETSVLDP Sbjct: 28 ASSTNLKDVLSNLIPKEQARIKTFKQQHGKTVVGQITVDMMYGGMRGMKGLVYETSVLDP 87 Query: 291 EEGIRFRGYSIPECQKVLPKSQGGEQPLPEGLFFLLLTGKIPTEEQVKWISKEWASRADL 470 +EGIRFRGYSIPECQK+LPK++GGE+PLPEGLF+LL+TG++PTEEQV W+S+EWA RA L Sbjct: 88 DEGIRFRGYSIPECQKMLPKAKGGEEPLPEGLFWLLVTGQMPTEEQVSWLSREWAKRAAL 147 Query: 471 PGHVVSMINNFPSSLHPMSQLVAASAALNSESKFVKAYHEGVSKSVYWETVYEDSMDLIA 650 P HVV+M++NFP++LHPMSQL AA ALNSES F +AY EG++++ YWE +YED MDLIA Sbjct: 148 PSHVVTMLDNFPTNLHPMSQLSAAITALNSESNFARAYAEGMNRAKYWELIYEDCMDLIA 207 Query: 651 KLSTVAAMIYRNLYRDGTSVGCIDNSKDWSENFVRMIGYEDPSFTELMRLYLTIHSDHEG 830 KL VAA IYRNLYR+G+S+G ID+ DWS NF M+GY DP FTELMRLYLTIHSDHEG Sbjct: 208 KLPCVAAKIYRNLYREGSSIGAIDSRLDWSHNFTNMLGYTDPQFTELMRLYLTIHSDHEG 267 Query: 831 GNVSAHTCHLVGSALSDPYLSYAAAMCGLAGPLHGLANQEVLLWVQKMIEVIGTKPTDKD 1010 GNVSAHT HLVGSALSDPYLS+AAAM GLAGPLHGLANQEVL+W+ ++ + +G +D+ Sbjct: 268 GNVSAHTSHLVGSALSDPYLSFAAAMNGLAGPLHGLANQEVLVWLTQLQKEVGKDVSDEK 327 Query: 1011 IVEFIEKTLKAGQVVPGYGHAVLRKTDPRYMCQREFALKHLPDYPLFKVVSQLYQIVPDV 1190 + ++I TL +G+VVPGYGHAVLRKTDPRY CQREFALKHLP P+FK+V+QLY+IVP++ Sbjct: 328 LRDYIWNTLNSGRVVPGYGHAVLRKTDPRYSCQREFALKHLPKDPMFKLVAQLYKIVPNI 387 Query: 1191 LGKKSKIANPFPNVDAHSGVLLQYYGMTETNYYTVLFGISRALGILSSLIWDRALGLPIE 1370 L ++ K NP+PNVDAHSGVLLQYYGMTE NYYTVLFG+SRALG+L+ LIW RALG P+E Sbjct: 388 LLEQGKAKNPWPNVDAHSGVLLQYYGMTEMNYYTVLFGVSRALGVLAQLIWSRALGFPLE 447 Query: 1371 RPKSFSTDKLIELV 1412 RPKS STD L++ V Sbjct: 448 RPKSMSTDGLMKFV 461
>sp|P23007|CISY_CHICK Citrate synthase, mitochondrial Length = 433 Score = 626 bits (1615), Expect = e-179 Identities = 302/433 (69%), Positives = 357/433 (82%) Frame = +3 Query: 111 ASTVNLKDILADTITKEQKKVSEFRKQHGETKVGEVTINMMYGGMRGIKGLVTETSVLDP 290 AS+ NLKD+LA I KEQ ++ FR+QHG T +G++T++M YGGMRG+KGLV ETSVLDP Sbjct: 1 ASSTNLKDVLAALIPKEQARIKTFRQQHGGTALGQITVDMSYGGMRGMKGLVYETSVLDP 60 Query: 291 EEGIRFRGYSIPECQKVLPKSQGGEQPLPEGLFFLLLTGKIPTEEQVKWISKEWASRADL 470 +EGIRFRG+SIPECQK+LPK G +PLPEGLF+LL+TG+IPT QV W+SKEWA RA L Sbjct: 61 DEGIRFRGFSIPECQKLLPKGGXGGEPLPEGLFWLLVTGQIPTGAQVSWLSKEWAKRAAL 120 Query: 471 PGHVVSMINNFPSSLHPMSQLVAASAALNSESKFVKAYHEGVSKSVYWETVYEDSMDLIA 650 P HVV+M++NFP++LHPMSQL AA ALNSES F +AY EG+ ++ YWE VYE +MDLIA Sbjct: 121 PSHVVTMLDNFPTNLHPMSQLSAAITALNSESNFARAYAEGILRTKYWEMVYESAMDLIA 180 Query: 651 KLSTVAAMIYRNLYRDGTSVGCIDNSKDWSENFVRMIGYEDPSFTELMRLYLTIHSDHEG 830 KL VAA IYRNLYR G+S+G ID+ DWS NF M+GY D FTELMRLYLTIHSDHEG Sbjct: 181 KLPCVAAKIYRNLYRAGSSIGAIDSKLDWSHNFTNMLGYTDAQFTELMRLYLTIHSDHEG 240 Query: 831 GNVSAHTCHLVGSALSDPYLSYAAAMCGLAGPLHGLANQEVLLWVQKMIEVIGTKPTDKD 1010 GNVSAHT HLVGSALSDPYLS+AAAM GLAGPLHGLANQEVL W+ ++ + D Sbjct: 241 GNVSAHTSHLVGSALSDPYLSFAAAMNGLAGPLHGLANQEVLGWLAQLQKAXXXAGADAS 300 Query: 1011 IVEFIEKTLKAGQVVPGYGHAVLRKTDPRYMCQREFALKHLPDYPLFKVVSQLYQIVPDV 1190 + ++I TL +G+VVPGYGHAVLRKTDPRY CQREFALKHLP P+FK+V+QLY+IVP+V Sbjct: 301 LRDYIWNTLNSGRVVPGYGHAVLRKTDPRYTCQREFALKHLPGDPMFKLVAQLYKIVPNV 360 Query: 1191 LGKKSKIANPFPNVDAHSGVLLQYYGMTETNYYTVLFGISRALGILSSLIWDRALGLPIE 1370 L ++ ANP+PNVDAHSGVLLQYYGMTE NYYTVLFG+SRALG+L+ LIW RALG P+E Sbjct: 361 LLEQGAAANPWPNVDAHSGVLLQYYGMTEMNYYTVLFGVSRALGVLAQLIWSRALGFPLE 420 Query: 1371 RPKSFSTDKLIEL 1409 RPKS STD LI L Sbjct: 421 RPKSMSTDGLIAL 433
>sp|P34575|CISY_CAEEL Probable citrate synthase, mitochondrial precursor Length = 468 Score = 606 bits (1563), Expect = e-173 Identities = 292/431 (67%), Positives = 354/431 (82%) Frame = +3 Query: 117 TVNLKDILADTITKEQKKVSEFRKQHGETKVGEVTINMMYGGMRGIKGLVTETSVLDPEE 296 + NLK++L+ I KV FR +HG T V V I+M+YGGMR +KG+VTETSVLDPEE Sbjct: 32 STNLKEVLSKKIPAHNAKVKSFRTEHGSTVVQNVNIDMIYGGMRSMKGMVTETSVLDPEE 91 Query: 297 GIRFRGYSIPECQKVLPKSQGGEQPLPEGLFFLLLTGKIPTEEQVKWISKEWASRADLPG 476 GIRFRGYSIPECQK+LPK++GGE+PLPE +++LL TG +P+E Q I+KEW +RADLP Sbjct: 92 GIRFRGYSIPECQKLLPKAKGGEEPLPEAIWWLLCTGDVPSEAQTAAITKEWNARADLPT 151 Query: 477 HVVSMINNFPSSLHPMSQLVAASAALNSESKFVKAYHEGVSKSVYWETVYEDSMDLIAKL 656 HVV M++NFP +LHPM+Q +AA AALN+ESKF AY GV+K+ YWE YEDSMDL+AKL Sbjct: 152 HVVRMLDNFPDNLHPMAQFIAAIAALNNESKFAGAYARGVAKASYWEYAYEDSMDLLAKL 211 Query: 657 STVAAMIYRNLYRDGTSVGCIDNSKDWSENFVRMIGYEDPSFTELMRLYLTIHSDHEGGN 836 TVAA+IYRNLYRDG++V ID KDWS NF M+GY+DP F ELMRLYL IHSDHEGGN Sbjct: 212 PTVAAIIYRNLYRDGSAVSVIDPKKDWSANFSSMLGYDDPLFAELMRLYLVIHSDHEGGN 271 Query: 837 VSAHTCHLVGSALSDPYLSYAAAMCGLAGPLHGLANQEVLLWVQKMIEVIGTKPTDKDIV 1016 VSAHT HLVGSALSDPYLS++AAM GLAGPLHGLANQEVL+++ K++ IG T++ + Sbjct: 272 VSAHTSHLVGSALSDPYLSFSAAMAGLAGPLHGLANQEVLVFLNKIVGEIGFNYTEEQLK 331 Query: 1017 EFIEKTLKAGQVVPGYGHAVLRKTDPRYMCQREFALKHLPDYPLFKVVSQLYQIVPDVLG 1196 E++ K LK+GQVVPGYGHAVLRKTDPRY CQREFALKHLP+ LFK+VS LY+I P +L Sbjct: 332 EWVWKHLKSGQVVPGYGHAVLRKTDPRYECQREFALKHLPNDDLFKLVSTLYKITPGILL 391 Query: 1197 KKSKIANPFPNVDAHSGVLLQYYGMTETNYYTVLFGISRALGILSSLIWDRALGLPIERP 1376 ++ K NP+PNVD+HSGVLLQY+GMTE ++YTVLFG+SRALG LS LIW R +GLP+ERP Sbjct: 392 EQGKAKNPWPNVDSHSGVLLQYFGMTEMSFYTVLFGVSRALGCLSQLIWARGMGLPLERP 451 Query: 1377 KSFSTDKLIEL 1409 KS STD LI+L Sbjct: 452 KSHSTDGLIKL 462
>sp|P51044|CISY_ASPNG Citrate synthase, mitochondrial precursor Length = 475 Score = 545 bits (1403), Expect = e-154 Identities = 273/436 (62%), Positives = 332/436 (76%), Gaps = 1/436 (0%) Frame = +3 Query: 90 GIRNVRFASTVNLKDILADTITKEQKKVSEFRKQHGETKVGEVTINMMYGGMRGIKGLVT 269 G+R +LK+ A+ + E +KV + RK+HG +GEVT++ YGG RG+K LV Sbjct: 30 GLRCYSTGKAKSLKETFAEKLPAELEKVKKLRKEHGSKVIGEVTLDQAYGGARGVKCLVW 89 Query: 270 ETSVLDPEEGIRFRGYSIPECQKVLPKSQGGEQPLPEGLFFLLLTGKIPTEEQVKWISKE 449 E SVLD EEGIRFRG +IPECQ++LPK+ GG++PLPEGLF+LLLTG+IPTE+QV+ +S E Sbjct: 90 EGSVLDSEEGIRFRGRTIPECQELLPKAPGGQEPLPEGLFWLLLTGEIPTEQQVRDLSAE 149 Query: 450 WASRADLPGHVVSMINNFPSSLHPMSQLVAASAALNSESKFVKAYHEGVSKSVYWETVYE 629 WA+R+DLP + +I+ PS+LHPMSQ A AL ES F KAY +G++K YW +E Sbjct: 150 WAARSDLPKFIEELIDRCPSTLHPMSQFSLAVTALEHESAFAKAYAKGINKKDYWNYTFE 209 Query: 630 DSMDLIAKLSTVAAMIYRNLYRDGTSVGCIDNSKDWSENFVRMIGYEDPS-FTELMRLYL 806 DSMDLIAKL T+AA IYRN+++DG V I KD+S N +GY D + F ELMRLYL Sbjct: 210 DSMDLIAKLPTIAAKIYRNVFKDG-KVAPIQKDKDYSYNLANQLGYGDNNDFVELMRLYL 268 Query: 807 TIHSDHEGGNVSAHTCHLVGSALSDPYLSYAAAMCGLAGPLHGLANQEVLLWVQKMIEVI 986 TIHSDHEGGNVSAHT HLVGSALS P LS AA + GLAGPLHGLANQEVL W+ KM I Sbjct: 269 TIHSDHEGGNVSAHTTHLVGSALSSPMLSLAAGLNGLAGPLHGLANQEVLNWLTKMKAAI 328 Query: 987 GTKPTDKDIVEFIEKTLKAGQVVPGYGHAVLRKTDPRYMCQREFALKHLPDYPLFKVVSQ 1166 G +D+ I ++ TL AGQVVPGYGHAVLRKTDPRY+ QREFAL+ LPD P+FK+VSQ Sbjct: 329 GNDLSDEAIKNYLWSTLNAGQVVPGYGHAVLRKTDPRYVSQREFALRKLPDDPMFKLVSQ 388 Query: 1167 LYQIVPDVLGKKSKIANPFPNVDAHSGVLLQYYGMTETNYYTVLFGISRALGILSSLIWD 1346 +Y+I P VL + K NP+PNVDAHSGVLLQYYG+TE NYYTVLFG+SRALG+L LI D Sbjct: 389 VYKIAPGVLTEHGKTKNPYPNVDAHSGVLLQYYGLTEANYYTVLFGVSRALGVLPQLIID 448 Query: 1347 RALGLPIERPKSFSTD 1394 RALG PIERPKS+ST+ Sbjct: 449 RALGAPIERPKSYSTE 464
>sp|O00098|CISY_EMENI Citrate synthase, mitochondrial precursor Length = 474 Score = 544 bits (1402), Expect = e-154 Identities = 271/442 (61%), Positives = 335/442 (75%), Gaps = 1/442 (0%) Frame = +3 Query: 90 GIRNVRFASTVNLKDILADTITKEQKKVSEFRKQHGETKVGEVTINMMYGGMRGIKGLVT 269 G+R T +LK+ AD + E +KV + RK+HG +GE+T++ YGG RG+K LV Sbjct: 30 GLRCYSTGKTKSLKETFADKLPGELEKVKKLRKEHGNKVIGELTLDQAYGGARGVKCLVW 89 Query: 270 ETSVLDPEEGIRFRGYSIPECQKVLPKSQGGEQPLPEGLFFLLLTGKIPTEEQVKWISKE 449 E SVLD EEGIRFRG +IPECQK+LPK+ GGE+PLPEGLF+LLLTG++P+E+QV+ +S E Sbjct: 90 EGSVLDSEEGIRFRGLTIPECQKLLPKAPGGEEPLPEGLFWLLLTGEVPSEQQVRDLSAE 149 Query: 450 WASRADLPGHVVSMINNFPSSLHPMSQLVAASAALNSESKFVKAYHEGVSKSVYWETVYE 629 WA+R+DLP + +I+ PS+LHPM+Q A AL ES F KA +G++K YW +E Sbjct: 150 WAARSDLPKFIEELIDRVPSTLHPMAQFSLAVTALEHESAFAKATAKGINKKEYWHYTFE 209 Query: 630 DSMDLIAKLSTVAAMIYRNLYRDGTSVGCIDNSKDWSENFVRMIGYED-PSFTELMRLYL 806 DSMDLIAKL T+AA IYRN+++DG V I KD+S N +G+ D F ELMRLYL Sbjct: 210 DSMDLIAKLPTIAAKIYRNVFKDG-KVAPIQKDKDYSYNLANQLGFADNKDFVELMRLYL 268 Query: 807 TIHSDHEGGNVSAHTCHLVGSALSDPYLSYAAAMCGLAGPLHGLANQEVLLWVQKMIEVI 986 TIHSDHEGGNVSAHT HLVGSALS P LS AA + GLAGPLHGLANQEVL W+ +M +V+ Sbjct: 269 TIHSDHEGGNVSAHTTHLVGSALSSPMLSLAAGLNGLAGPLHGLANQEVLNWLTEMKKVV 328 Query: 987 GTKPTDKDIVEFIEKTLKAGQVVPGYGHAVLRKTDPRYMCQREFALKHLPDYPLFKVVSQ 1166 G +D+ I +++ TL AG+VVPGYGHAVLRKTDPRY QREFAL+ LPD P+FK+VSQ Sbjct: 329 GNDLSDQSIKDYLWSTLNAGRVVPGYGHAVLRKTDPRYTSQREFALRKLPDDPMFKLVSQ 388 Query: 1167 LYQIVPDVLGKKSKIANPFPNVDAHSGVLLQYYGMTETNYYTVLFGISRALGILSSLIWD 1346 +Y+I P VL + K NP+PNVDAHSGVLLQYYG+TE NYYTVLFG+SRALG+L LI D Sbjct: 389 VYKIAPGVLTEHGKTKNPYPNVDAHSGVLLQYYGLTEANYYTVLFGVSRALGVLPQLIID 448 Query: 1347 RALGLPIERPKSFSTDKLIELV 1412 RA G PIERPKSFST+ +LV Sbjct: 449 RAFGAPIERPKSFSTEAYAKLV 470
>sp|P00890|CISY1_YEAST Citrate synthase, mitochondrial precursor Length = 479 Score = 537 bits (1383), Expect = e-152 Identities = 261/448 (58%), Positives = 339/448 (75%) Frame = +3 Query: 75 VFLSCGIRNVRFASTVNLKDILADTITKEQKKVSEFRKQHGETKVGEVTINMMYGGMRGI 254 +F R+ AS LK+ A+ I + +++ +F+K+HG+T +GEV + YGGMRGI Sbjct: 28 LFALLNARHYSSASEQTLKERFAEIIPAKAEEIKKFKKEHGKTVIGEVLLEQAYGGMRGI 87 Query: 255 KGLVTETSVLDPEEGIRFRGYSIPECQKVLPKSQGGEQPLPEGLFFLLLTGKIPTEEQVK 434 KGLV E SVLDPEEGIRFRG +IPE Q+ LPK++G +PLPE LF+LLLTG+IPT+ QVK Sbjct: 88 KGLVWEGSVLDPEEGIRFRGRTIPEIQRELPKAEGSTEPLPEALFWLLLTGEIPTDAQVK 147 Query: 435 WISKEWASRADLPGHVVSMINNFPSSLHPMSQLVAASAALNSESKFVKAYHEGVSKSVYW 614 +S + A+R+++P HV+ ++++ P LHPM+Q A AL SESKF KAY +GVSK YW Sbjct: 148 ALSADLAARSEIPEHVIQLLDSLPKDLHPMAQFSIAVTALESESKFAKAYAQGVSKKEYW 207 Query: 615 ETVYEDSMDLIAKLSTVAAMIYRNLYRDGTSVGCIDNSKDWSENFVRMIGYEDPSFTELM 794 +EDS+DL+ KL +A+ IYRN+++DG + D + D+ +N +++GYE+ F +LM Sbjct: 208 SYTFEDSLDLLGKLPVIASKIYRNVFKDG-KITSTDPNADYGKNLAQLLGYENKDFIDLM 266 Query: 795 RLYLTIHSDHEGGNVSAHTCHLVGSALSDPYLSYAAAMCGLAGPLHGLANQEVLLWVQKM 974 RLYLTIHSDHEGGNVSAHT HLVGSALS PYLS AA + GLAGPLHG ANQEVL W+ K+ Sbjct: 267 RLYLTIHSDHEGGNVSAHTTHLVGSALSSPYLSLAAGLNGLAGPLHGRANQEVLEWLFKL 326 Query: 975 IEVIGTKPTDKDIVEFIEKTLKAGQVVPGYGHAVLRKTDPRYMCQREFALKHLPDYPLFK 1154 E + + + I +++ TL AG+VVPGYGHAVLRKTDPRY QREFALKH PDY LFK Sbjct: 327 REEVKGDYSKETIEKYLWDTLNAGRVVPGYGHAVLRKTDPRYTAQREFALKHFPDYELFK 386 Query: 1155 VVSQLYQIVPDVLGKKSKIANPFPNVDAHSGVLLQYYGMTETNYYTVLFGISRALGILSS 1334 +VS +Y++ P VL K K NP+PNVD+HSGVLLQYYG+TE ++YTVLFG++RA+G+L Sbjct: 387 LVSTIYEVAPGVLTKHGKTKNPWPNVDSHSGVLLQYYGLTEASFYTVLFGVARAIGVLPQ 446 Query: 1335 LIWDRALGLPIERPKSFSTDKLIELVRK 1418 LI DRA+G PIERPKSFST+K ELV+K Sbjct: 447 LIIDRAVGAPIERPKSFSTEKYKELVKK 474
>sp|O80433|CISY_DAUCA Citrate synthase, mitochondrial precursor Length = 472 Score = 536 bits (1381), Expect = e-152 Identities = 255/437 (58%), Positives = 337/437 (77%) Frame = +3 Query: 111 ASTVNLKDILADTITKEQKKVSEFRKQHGETKVGEVTINMMYGGMRGIKGLVTETSVLDP 290 AS ++L+ L + I ++Q+++ + + +HG+ ++G +T++M+ GGMRG+ GL+ ETS+LDP Sbjct: 36 ASDLDLRSQLKELIPEQQERIKKLKAEHGKVQLGNITVDMVLGGMRGMTGLLWETSLLDP 95 Query: 291 EEGIRFRGYSIPECQKVLPKSQGGEQPLPEGLFFLLLTGKIPTEEQVKWISKEWASRADL 470 EEGIRFRG SIPECQK+LP ++ G +PLPEGL +LLLTGK+PT+EQV +S E SRA + Sbjct: 96 EEGIRFRGLSIPECQKLLPGAKPGGEPLPEGLLWLLLTGKVPTKEQVDALSAELRSRAAV 155 Query: 471 PGHVVSMINNFPSSLHPMSQLVAASAALNSESKFVKAYHEGVSKSVYWETVYEDSMDLIA 650 P HV I+ P + HPM+Q AL +S+F KAY +G+ K+ YWE YEDS+ LIA Sbjct: 156 PEHVYKTIDALPVTAHPMTQFATGVMALQVQSEFQKAYEKGIHKTKYWEPTYEDSITLIA 215 Query: 651 KLSTVAAMIYRNLYRDGTSVGCIDNSKDWSENFVRMIGYEDPSFTELMRLYLTIHSDHEG 830 +L VAA IYR +Y++G S+ D+S D+ NF M+GY+ PS ELMRLY+TIH+DHEG Sbjct: 216 QLPVVAAYIYRRMYKNGQSIST-DDSLDYGANFAHMLGYDSPSMQELMRLYVTIHTDHEG 274 Query: 831 GNVSAHTCHLVGSALSDPYLSYAAAMCGLAGPLHGLANQEVLLWVQKMIEVIGTKPTDKD 1010 GNVSAHT HLV SALSDPYLS+AAA+ GLAGPLHGLANQEVLLW++ ++ G T + Sbjct: 275 GNVSAHTGHLVASALSDPYLSFAAALNGLAGPLHGLANQEVLLWIKSVVSECGENVTKEQ 334 Query: 1011 IVEFIEKTLKAGQVVPGYGHAVLRKTDPRYMCQREFALKHLPDYPLFKVVSQLYQIVPDV 1190 + ++I KTL +G+VVPGYGH VLR TDPRY+CQREFALKHLPD PLF++VS L+++VP + Sbjct: 335 LKDYIWKTLNSGKVVPGYGHGVLRNTDPRYICQREFALKHLPDDPLFQLVSNLFEVVPPI 394 Query: 1191 LGKKSKIANPFPNVDAHSGVLLQYYGMTETNYYTVLFGISRALGILSSLIWDRALGLPIE 1370 L + K+ NP+PNVDAHSGVLL +YG+TE YYTVLFG+SRA+GI S L+WDRALGLP+E Sbjct: 395 LTELGKVKNPWPNVDAHSGVLLNHYGLTEARYYTVLFGVSRAIGICSQLVWDRALGLPLE 454 Query: 1371 RPKSFSTDKLIELVRKS 1421 RPKS + + L +KS Sbjct: 455 RPKSVTMEWLENHCKKS 471
>sp|P20115|CISY4_ARATH Citrate synthase 4, mitochondrial precursor Length = 474 Score = 536 bits (1380), Expect = e-152 Identities = 256/442 (57%), Positives = 344/442 (77%), Gaps = 2/442 (0%) Frame = +3 Query: 81 LSCGIRNVRFAST--VNLKDILADTITKEQKKVSEFRKQHGETKVGEVTINMMYGGMRGI 254 LS +R ++ S+ ++LK L + I ++Q ++ + + +HG+ ++G +T++M+ GGMRG+ Sbjct: 24 LSNSVRWIQMQSSTDLDLKSQLQELIPEQQDRLKKLKSEHGKVQLGNITVDMVIGGMRGM 83 Query: 255 KGLVTETSVLDPEEGIRFRGYSIPECQKVLPKSQGGEQPLPEGLFFLLLTGKIPTEEQVK 434 GL+ ETS+LDPEEGIRFRG SIPECQKVLP +Q G +PLPEGL +LLLTGK+P++EQV+ Sbjct: 84 TGLLWETSLLDPEEGIRFRGLSIPECQKVLPTAQSGAEPLPEGLLWLLLTGKVPSKEQVE 143 Query: 435 WISKEWASRADLPGHVVSMINNFPSSLHPMSQLVAASAALNSESKFVKAYHEGVSKSVYW 614 +SK+ A+RA +P +V + I+ PS+ HPM+Q + AL +S+F KAY G+ KS +W Sbjct: 144 ALSKDLANRAAVPDYVYNAIDALPSTAHPMTQFASGVMALQVQSEFQKAYENGIHKSKFW 203 Query: 615 ETVYEDSMDLIAKLSTVAAMIYRNLYRDGTSVGCIDNSKDWSENFVRMIGYEDPSFTELM 794 E YED ++LIA++ VAA +YR +Y++G S+ D S D+ NF M+G++D ELM Sbjct: 204 EPTYEDCLNLIARVPVVAAYVYRRMYKNGDSIPS-DKSLDYGANFSHMLGFDDEKVKELM 262 Query: 795 RLYLTIHSDHEGGNVSAHTCHLVGSALSDPYLSYAAAMCGLAGPLHGLANQEVLLWVQKM 974 RLY+TIHSDHEGGNVSAHT HLVGSALSDPYLS+AAA+ GLAGPLHGLANQEVLLW++ + Sbjct: 263 RLYITIHSDHEGGNVSAHTGHLVGSALSDPYLSFAAALNGLAGPLHGLANQEVLLWIKSV 322 Query: 975 IEVIGTKPTDKDIVEFIEKTLKAGQVVPGYGHAVLRKTDPRYMCQREFALKHLPDYPLFK 1154 +E G + + + E++ KTL +G+V+PGYGH VLR TDPRY+CQREFALKHLPD PLF+ Sbjct: 323 VEECGEDISKEQLKEYVWKTLNSGKVIPGYGHGVLRNTDPRYVCQREFALKHLPDDPLFQ 382 Query: 1155 VVSQLYQIVPDVLGKKSKIANPFPNVDAHSGVLLQYYGMTETNYYTVLFGISRALGILSS 1334 +VS+LY++VP VL + K+ NP+PNVDAHSGVLL +YG+TE YYTVLFG+SR+LGI S Sbjct: 383 LVSKLYEVVPPVLTELGKVKNPWPNVDAHSGVLLNHYGLTEARYYTVLFGVSRSLGICSQ 442 Query: 1335 LIWDRALGLPIERPKSFSTDKL 1400 LIWDRALGL +ERPKS + D L Sbjct: 443 LIWDRALGLALERPKSVTMDWL 464
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 186,385,082
Number of Sequences: 369166
Number of extensions: 4015482
Number of successful extensions: 10676
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9775
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10469
length of database: 68,354,980
effective HSP length: 116
effective length of database: 46,925,720
effective search space used: 19849579560
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail