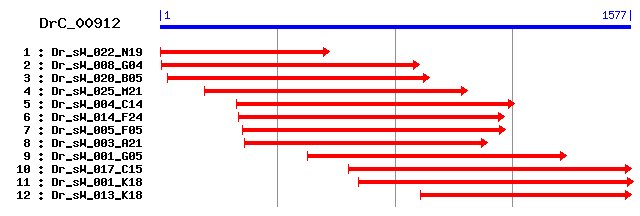
DrC_00912
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00912 (1577 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P79287|MMP20_PIG Matrix metalloproteinase-20 precursor (... 137 9e-32 sp|O60882|MMP20_HUMAN Matrix metalloproteinase-20 precursor... 137 1e-31 sp|O18767|MMP20_BOVIN Matrix metalloproteinase-20 precursor... 137 1e-31 sp|Q9JHI0|MMP19_MOUSE Matrix metalloproteinase-19 precursor... 135 4e-31 sp|P21692|MMP1_PIG Interstitial collagenase precursor (Matr... 131 5e-30 sp|P57748|MMP20_MOUSE Matrix metalloproteinase-20 precursor... 131 6e-30 sp|O62806|MMP13_RABIT Collagenase 3 precursor (Matrix metal... 130 1e-29 sp|P22894|MMP8_HUMAN Neutrophil collagenase precursor (Matr... 129 2e-29 sp|P28053|MMP1_BOVIN Interstitial collagenase precursor (Ma... 128 4e-29 sp|P45452|MMP13_HUMAN Collagenase 3 precursor (Matrix metal... 128 4e-29
>sp|P79287|MMP20_PIG Matrix metalloproteinase-20 precursor (MMP-20) (Enamel metalloproteinase) (Enamelysin) Length = 483 Score = 137 bits (345), Expect = 9e-32 Identities = 114/364 (31%), Positives = 167/364 (45%), Gaps = 9/364 (2%) Frame = +1 Query: 130 VKNIQKMAGLPTDGKINKELLAHVGKSRCGGKDVKPKNKRYNAANSKWDKGT-TYRFYIH 306 +K +Q GL GK+++ + + + RCG DV N R KW K T TYR Sbjct: 72 IKELQAFFGLRVTGKLDRTTMDVIKRPRCGVPDVA--NYRLFPGEPKWKKNTLTYRI--- 126 Query: 307 DERHPTTMTLGEIQRALMIGLSYWSTAGDIYFAKVSNKQGSMFSLSFADAEHVQYDVHNN 486 ++ +MT E+ +A+ + L WS+A + F +V+ + + +SF +H Sbjct: 127 -SKYTPSMTPAEVDKAMEMALQAWSSAVPLSFVRVNAGEADIM-ISFETGDH-------- 176 Query: 487 FVESRDPYPLDNVPGGVLAHAFFPRNG---NLHFDSAEEWTEGKEDGINLRLVAAHEMGH 657 D YP D P G LAHAF P G + HFD+AE+WT G +G NL VAAHE GH Sbjct: 177 ----GDSYPFDG-PRGTLAHAFAPGEGLGGDTHFDNAEKWTMGM-NGFNLFTVAAHEFGH 230 Query: 658 AIGLFHSNDPNALMAPWYGGYISENDFKLPDDDSQGAAFIYG---TKNGKVFVRSNKKRE 828 A+GL HS DP+ALM P Y Y + F LP DD +G +YG T GK V Sbjct: 231 ALGLAHSTDPSALMYPTY-KYQNPYGFHLPKDDVKGIQALYGPRKTFTGKPTVPHGPPHN 289 Query: 829 QSANEKELCNITNINSAFQYSDDNIYVVTDKNKVFRIIHSNNSFRVDASFKTLQLPNQLF 1008 S ++C+ ++ A + D+ R +H + R S T P + Sbjct: 290 PSL--PDICDSSSSFDAVTMLGKELLFFRDRIFWRRQVHLMSGIR--PSTITSSFPQLMS 345 Query: 1009 DSDALFYDYSGRRFVFFKGNRVFSANGVKLEN--LTISELNVTRPGGFQGRVTAAIVRDK 1182 + DA + FFKG + G +++ TI + R + R+ AA+ Sbjct: 346 NVDAAYEVADRGMAYFFKGPHYWITRGFQMQGPPRTIYDFGFPR---YVQRIDAAVHLKD 402 Query: 1183 TGKS 1194 T K+ Sbjct: 403 TQKT 406
>sp|O60882|MMP20_HUMAN Matrix metalloproteinase-20 precursor (MMP-20) (Enamel metalloproteinase) (Enamelysin) Length = 483 Score = 137 bits (344), Expect = 1e-31 Identities = 104/331 (31%), Positives = 157/331 (47%), Gaps = 7/331 (2%) Frame = +1 Query: 130 VKNIQKMAGLPTDGKINKELLAHVGKSRCGGKDVKPKNKRYNAANSKWDKGT-TYRFYIH 306 +K +Q GL GK+++ + + K RCG DV N R KW K T TYR Sbjct: 72 IKELQAFFGLQVTGKLDQTTMNVIKKPRCGVPDVA--NYRLFPGEPKWKKNTLTYRI--- 126 Query: 307 DERHPTTMTLGEIQRALMIGLSYWSTAGDIYFAKVSNKQGSMFSLSFADAEHVQYDVHNN 486 ++ +M+ E+ +A+ + L WS+A + F ++++ + + +SF + +H Sbjct: 127 -SKYTPSMSSVEVDKAVEMALQAWSSAVPLSFVRINSGEADIM-ISFENGDH-------- 176 Query: 487 FVESRDPYPLDNVPGGVLAHAFFPRNG---NLHFDSAEEWTEGKEDGINLRLVAAHEMGH 657 D YP D P G LAHAF P G + HFD+AE+WT G +G NL VAAHE GH Sbjct: 177 ----GDSYPFDG-PRGTLAHAFAPGEGLGGDTHFDNAEKWTMGT-NGFNLFTVAAHEFGH 230 Query: 658 AIGLFHSNDPNALMAPWYGGYISENDFKLPDDDSQGAAFIYGTKNGKVFVRS---NKKRE 828 A+GL HS DP+ALM P Y Y + F LP DD +G +YG + KVF+ Sbjct: 231 ALGLAHSTDPSALMYPTY-KYKNPYGFHLPKDDVKGIQALYGPR--KVFLGKPTLPHAPH 287 Query: 829 QSANEKELCNITNINSAFQYSDDNIYVVTDKNKVFRIIHSNNSFRVDASFKTLQLPNQLF 1008 + +LC+ ++ A + + D+ R +H R S T P + Sbjct: 288 HKPSIPDLCDSSSSFDAVTMLGKELLLFKDRIFWRRQVHLRTGIR--PSTITSSFPQLMS 345 Query: 1009 DSDALFYDYSGRRFVFFKGNRVFSANGVKLE 1101 + DA + FFKG + G +++ Sbjct: 346 NVDAAYEVAERGTAYFFKGPHYWITRGFQMQ 376
>sp|O18767|MMP20_BOVIN Matrix metalloproteinase-20 precursor (MMP-20) (Enamel metalloproteinase) (Enamelysin) Length = 481 Score = 137 bits (344), Expect = 1e-31 Identities = 105/331 (31%), Positives = 155/331 (46%), Gaps = 7/331 (2%) Frame = +1 Query: 130 VKNIQKMAGLPTDGKINKELLAHVGKSRCGGKDVKPKNKRYNAANSKWDKGT-TYRFYIH 306 +K +Q+ GL GK+++ + + + RCG DV N R KW K T TYR Sbjct: 70 IKELQEFFGLRVTGKLDRATMDVIKRPRCGVPDVA--NYRLFPGEPKWKKNTLTYRI--- 124 Query: 307 DERHPTTMTLGEIQRALMIGLSYWSTAGDIYFAKVSNKQGSMFSLSFADAEHVQYDVHNN 486 ++ +MT E+ RA+ + L WS+A + F +++ + + +SF +H Sbjct: 125 -SKYTPSMTPAEVDRAMEMALRAWSSAVPLNFVRINAGEADIM-ISFETGDH-------- 174 Query: 487 FVESRDPYPLDNVPGGVLAHAFFPRNG---NLHFDSAEEWTEGKEDGINLRLVAAHEMGH 657 D YP D P G LAHAF P G + HFD+AE+WT G +G NL VAAHE GH Sbjct: 175 ----GDSYPFDG-PRGTLAHAFAPGEGLGGDTHFDNAEKWTMGT-NGFNLFTVAAHEFGH 228 Query: 658 AIGLFHSNDPNALMAPWYGGYISENDFKLPDDDSQGAAFIYGTK---NGKVFVRSNKKRE 828 A+GL HS DP+ALM P Y Y + F+LP DD +G +YG + +GK Sbjct: 229 ALGLAHSTDPSALMFPTY-KYQNPYGFRLPKDDVKGIQALYGPRRAFSGKPTAPHGPPHN 287 Query: 829 QSANEKELCNITNINSAFQYSDDNIYVVTDKNKVFRIIHSNNSFRVDASFKTLQLPNQLF 1008 S +LC+ A + + D+ R +H + R S T P + Sbjct: 288 PSI--PDLCDSNLSFDAVTMLGKELLLFRDRIFWRRQVHLMSGIR--PSTITSSFPQLMS 343 Query: 1009 DSDALFYDYSGRRFVFFKGNRVFSANGVKLE 1101 + DA + FFKG + G +++ Sbjct: 344 NVDAAYEVAERGTAYFFKGPHYWITRGFQMQ 374
>sp|Q9JHI0|MMP19_MOUSE Matrix metalloproteinase-19 precursor (MMP-19) (Matrix metalloproteinase RASI) Length = 527 Score = 135 bits (339), Expect = 4e-31 Identities = 105/354 (29%), Positives = 162/354 (45%), Gaps = 16/354 (4%) Frame = +1 Query: 64 YVEKFLSDFYDYNENTDRLNDAVKNIQKMAGLPTDGKINKELLAHVGKSRCGGKD-VKPK 240 Y++K L D+ + + +A++ Q+ +GLP G+++ A + + RCG +D K Sbjct: 38 YLQKPLEGADDFR--LEDITEALRTFQEASGLPISGQMDDATRARMKQPRCGLEDPFNQK 95 Query: 241 NKRYNAANSKWDKGTTYRFYIHDERHPTTMTLGEIQRALMIGLSYWSTAGDIYFAKVSNK 420 + +Y K T+R + P+T++L ++ AL YWS+ + F +V Sbjct: 96 SLKYLLLGHWRKKNLTFRIF----NVPSTLSLPRVRAALHQAFKYWSSVAPLTFREVKAG 151 Query: 421 QGSMFSLSFADAEHVQYDVHNNFVESRDPYPLDNVPGGVLAHAFFPRNGNLHFDSAEEWT 600 + LSF + + N F + PG VLAHA P G++HFD E WT Sbjct: 152 WADI-RLSFHGRQSLYCS--NTF----------DGPGKVLAHADIPELGSIHFDKDELWT 198 Query: 601 EGKEDGINLRLVAAHEMGHAIGLFHSNDPNALMAPWYGGYISENDFKLPDDDSQGAAFIY 780 EG G+NLR++AAHE+GHA+GL HS ALMAP Y GY + FKL DD G +Y Sbjct: 199 EGTYQGVNLRIIAAHEVGHALGLGHSRYTQALMAPVYAGY--QPFFKLHPDDVAGIQALY 256 Query: 781 GTKN---------------GKVFVRSNKKREQSANEKELCNITNINSAFQYSDDNIYVVT 915 G ++ V + + E + + + + D ++ VT Sbjct: 257 GKRSPETRDEEEETEMLTVSPVTAKPGPMPNPCSGEVDAMVLGPRGKTYAFKGDYVWTVT 316 Query: 916 DKNKVFRIIHSNNSFRVDASFKTLQLPNQLFDSDALFYDYSGRRFVFFKGNRVF 1077 D F++ A ++ LP L DA Y RR FFKGN+V+ Sbjct: 317 DSG-------PGPLFQISALWE--GLPGNL---DAAVYSPRTRRTHFFKGNKVW 358
>sp|P21692|MMP1_PIG Interstitial collagenase precursor (Matrix metalloproteinase-1) (MMP-1) [Contains: 18 kDa interstitial collagenase] Length = 469 Score = 131 bits (330), Expect = 5e-30 Identities = 120/405 (29%), Positives = 178/405 (43%), Gaps = 22/405 (5%) Frame = +1 Query: 46 ESSDEKYVEKFLSDFYDYNE---------NTDRLNDAVKNIQKMAGLPTDGKINKELLAH 198 + D + V+K+L ++Y+ N N+ + + +K +Q+ GL GK + E L Sbjct: 27 QEQDVEIVQKYLKNYYNLNSDGVPVEKKRNSGLVVEKLKQMQQFFGLKVTGKPDAETLNV 86 Query: 199 VGKSRCGGKDVKPKNKRYNAANSKWDKG-TTYRFYIHDERHPTTMTLGEIQRALMIGLSY 375 + + RCG DV N +W+ TYR E + ++ ++ RA+ Sbjct: 87 MKQPRCGVPDVA--EFVLTPGNPRWENTHLTYRI----ENYTPDLSREDVDRAIEKAFQL 140 Query: 376 WSTAGDIYFAKVSNKQGSMFSLSFADAEHVQYDVHNNFVESRDPYPLDNVPGGVLAHAFF 555 WS + F KVS Q + +SF +H RD P D PGG LAHAF Sbjct: 141 WSNVSPLTFTKVSEGQADIM-ISFVRGDH------------RDNSPFDG-PGGNLAHAFQ 186 Query: 556 PR---NGNLHFDSAEEWTEGKEDGINLRLVAAHEMGHAIGLFHSNDPNALMAPWYGGYIS 726 P G+ HFD E WT+ D NL VAAHE+GH++GL HS D ALM Y YI Sbjct: 187 PGPGIGGDAHFDEDERWTKNFRD-YNLYRVAAHELGHSLGLSHSTDIGALM---YPNYIY 242 Query: 727 ENDFKLPDDDSQGAAFIYGTKNGKVFVRSNKKREQSANEKELCNITNINSAFQYSDDNIY 906 D +L DD G IYG V + + ++ IT + + D Y Sbjct: 243 TGDVQLSQDDIDGIQAIYGPSENPVQPSGPQTPQVCDSKLTFDAITTLRGELMFFKDRFY 302 Query: 907 VVTDKNKVFRIIHSNNSF--RVDASFKTL---QLPNQLFDSDALFYDYSGRRFV-FFKGN 1068 + T NSF V+ +F ++ Q+PN L Y+ + R V FFKGN Sbjct: 303 MRT------------NSFYPEVELNFISVFWPQVPNGL----QAAYEIADRDEVRFFKGN 346 Query: 1069 RVFSANGVKLENLTISELNVTRPGGFQG---RVTAAIVRDKTGKS 1194 + ++ G + L ++ R GF + AA+ + TGK+ Sbjct: 347 KYWAVRGQDV--LYGYPKDIHRSFGFPSTVKNIDAAVFEEDTGKT 389
>sp|P57748|MMP20_MOUSE Matrix metalloproteinase-20 precursor (MMP-20) (Enamel metalloproteinase) (Enamelysin) Length = 482 Score = 131 bits (329), Expect = 6e-30 Identities = 104/331 (31%), Positives = 153/331 (46%), Gaps = 7/331 (2%) Frame = +1 Query: 130 VKNIQKMAGLPTDGKINKELLAHVGKSRCGGKDVKPKNKRYNAANSKWDKGT-TYRFYIH 306 +K +Q GL GK+++ + + K RCG DV N R KW K TYR Sbjct: 71 IKELQIFFGLKVTGKLDQNTMNVIKKPRCGVPDVA--NYRLFPGEPKWKKNILTYRI--- 125 Query: 307 DERHPTTMTLGEIQRALMIGLSYWSTAGDIYFAKVSNKQGSMFSLSFADAEHVQYDVHNN 486 ++ +M+ E+ +A+ + L WSTA + F ++++ + + +SF +H Sbjct: 126 -SKYTPSMSPTEVDKAIQMALHAWSTAVPLNFVRINSGEADIM-ISFETGDH-------- 175 Query: 487 FVESRDPYPLDNVPGGVLAHAFFPRNG---NLHFDSAEEWTEGKEDGINLRLVAAHEMGH 657 D YP D P G LAHAF P G + HFD+AE+WT G +G NL VAAHE GH Sbjct: 176 ----GDSYPFDG-PRGTLAHAFAPGEGLGGDTHFDNAEKWTMGT-NGFNLFTVAAHEFGH 229 Query: 658 AIGLFHSNDPNALMAPWYGGYISENDFKLPDDDSQGAAFIYGTKN---GKVFVRSNKKRE 828 A+GL HS DP+ALM P Y Y + F LP DD +G +YG + GK + + Sbjct: 230 ALGLGHSTDPSALMYPTY-KYQNPYRFHLPKDDVKGIQALYGPRKIFPGKPTMPHIPPHK 288 Query: 829 QSANEKELCNITNINSAFQYSDDNIYVVTDKNKVFRIIHSNNSFRVDASFKTLQLPNQLF 1008 S +LC+ ++ A + D+ R +H R S T P + Sbjct: 289 PSI--PDLCDSSSSFDAVTMLGKELLFFKDRIFWRRQVHLPTGIR--PSTITSSFPQLMS 344 Query: 1009 DSDALFYDYSGRRFVFFKGNRVFSANGVKLE 1101 + DA + FFKG + G ++ Sbjct: 345 NVDAAYEVAERGIAFFFKGPHYWVTRGFHMQ 375
>sp|O62806|MMP13_RABIT Collagenase 3 precursor (Matrix metalloproteinase-13) (MMP-13) Length = 471 Score = 130 bits (326), Expect = 1e-29 Identities = 114/396 (28%), Positives = 173/396 (43%), Gaps = 16/396 (4%) Frame = +1 Query: 55 DEKYVEKFLSDFYD--------YNENTDRLNDAVKNIQKMAGLPTDGKINKELLAHVGKS 210 D ++ E +L +Y + D ++ +Q GL GK++ LA + + Sbjct: 35 DFQFAESYLRSYYHPLNPAGILKKNAAGSMVDRLREMQSFFGLEVTGKLDDNTLAIMKQP 94 Query: 211 RCGGKDVKPKNKRYNAANSKWDK-GTTYRFYIHDERHPTTMTLGEIQRALMIGLSYWSTA 387 RCG DV N KW + TYR + +T E+++A WS Sbjct: 95 RCGVPDVGEYN--VFPRTLKWSQTNLTYRIV----NYTPDLTHSEVEKAFKKAFKVWSDV 148 Query: 388 GDIYFAKVSNKQGSMFSLSFADAEHVQYDVHNNFVESRDPYPLDNVPGGVLAHAFFPR-- 561 + F ++ N + +SF EH + YP D P G+LAHAF P Sbjct: 149 TPLNFTRIHNGTADIM-ISFGTKEHGDF------------YPFDG-PSGLLAHAFPPGPN 194 Query: 562 -NGNLHFDSAEEWTEGKEDGINLRLVAAHEMGHAIGLFHSNDPNALMAPWYGGYISENDF 738 G+ HFD E WT + G NL LVAAHE GH++GL HS DP ALM P Y Y ++ F Sbjct: 195 YGGDAHFDDDETWTSSSK-GYNLFLVAAHEFGHSLGLDHSKDPGALMFPIY-TYTGKSHF 252 Query: 739 KLPDDDSQGAAFIYGTKNGKVFVRSNKKREQSANEKELCNITNINSAFQYSDDNIYVVTD 918 LPDDD QG +YG + + K ++ L IT++ D + Sbjct: 253 MLPDDDVQGIQSLYGPGDEDPNPKHPKTPDKCDPSLSLDAITSLRGETMIFKDRFFWRLH 312 Query: 919 KNKVFRIIHSNNSFRVDASFKTLQLPNQLFDSDALFYDYSGRRFVF-FKGNRVFSANGVK 1095 +V + SF +LPN++ DA Y++ R +F F+G + ++ NG Sbjct: 313 PQQVDAELFLTKSF-------WPELPNRI---DAA-YEHPARDLIFIFRGKKFWAPNGYD 361 Query: 1096 LEN---LTISELNVTRPGGFQGRVTAAIVRDKTGKS 1194 + +SEL R +++AA+ + TGK+ Sbjct: 362 ILEGYPQKLSELGFpre---VKKISAAVHFEDTGKT 394
>sp|P22894|MMP8_HUMAN Neutrophil collagenase precursor (Matrix metalloproteinase-8) (MMP-8) (PMNL collagenase) (PMNL-CL) Length = 467 Score = 129 bits (324), Expect = 2e-29 Identities = 114/403 (28%), Positives = 174/403 (43%), Gaps = 14/403 (3%) Frame = +1 Query: 46 ESSDEKYVEKFLSDFYDYNEN---------TDRLNDAVKNIQKMAGLPTDGKINKELLAH 198 + + K V+ +L FY N T+ + + +K +Q+ GL GK N+E L Sbjct: 26 KEKNTKTVQDYLEKFYQLPSNQYQSTRKNGTNVIVEKLKEMQRFFGLNVTGKPNEETLDM 85 Query: 199 VGKSRCGGKDVKPKNKRYNAANSKWDK-GTTYRFYIHDERHPTTMTLGEIQRALMIGLSY 375 + K RCG D N KW++ TYR + ++ E++RA+ Sbjct: 86 MKKPRCGVPD--SGGFMLTPGNPKWERTNLTYRI----RNYTPQLSEAEVERAIKDAFEL 139 Query: 376 WSTAGDIYFAKVSNKQGSMFSLSFADAEHVQYDVHNNFVESRDPYPLDNVPGGVLAHAFF 555 WS A + F ++S + + +++F +H D P D P G+LAHAF Sbjct: 140 WSVASPLIFTRISQGEADI-NIAFYQRDH------------GDNSPFDG-PNGILAHAFQ 185 Query: 556 PRNG---NLHFDSAEEWTEGKEDGINLRLVAAHEMGHAIGLFHSNDPNALMAPWYGGYIS 726 P G + HFD+ E WT + NL LVAAHE GH++GL HS+DP ALM P Y + Sbjct: 186 PGQGIGGDAHFDAEETWTNTSAN-YNLFLVAAHEFGHSLGLAHSSDPGALMYPNY-AFRE 243 Query: 727 ENDFKLPDDDSQGAAFIYGTKNGKVFVRSNKKREQSANEKELCNITNINSAFQYSDDNIY 906 +++ LP DD G IYG + SN + + + C+ + A I Sbjct: 244 TSNYSLPQDDIDGIQAIYG-------LSSNPIQPTGPSTPKPCDPSLTFDAITTLRGEIL 296 Query: 907 VVTDKNKVFRIIHSNNSFRVDASFKTLQLPNQLFDSDALFYDYSGRRFVFFKGNRVFSAN 1086 K++ F H RV+ +F +L P+ A + D+ FKGN+ ++ + Sbjct: 297 FF--KDRYFWRRHPQLQ-RVEMNFISLFWPSLPTGIQAAYEDFDRDLIFLFKGNQYWALS 353 Query: 1087 GVK-LENLTISELNVTRPGGFQGRVTAAIVRDKTGKSRTLKFW 1212 G L+ N P Q A R KT +FW Sbjct: 354 GYDILQGYPKDISNYGFPSSVQAIDAAVFYRSKTYFFVNDQFW 396
>sp|P28053|MMP1_BOVIN Interstitial collagenase precursor (Matrix metalloproteinase-1) (MMP-1) (Fibroblast collagenase) Length = 469 Score = 128 bits (322), Expect = 4e-29 Identities = 121/400 (30%), Positives = 172/400 (43%), Gaps = 17/400 (4%) Frame = +1 Query: 46 ESSDEKYVEKFLSDFYDYNENTDR---------LNDAVKNIQKMAGLPTDGKINKELLAH 198 + D + V+K+L ++Y+ N N + + + +K +QK GL GK + E L Sbjct: 27 QEQDVETVKKYLENYYNLNSNGKKVERQRNGGLITEKLKQMQKFFGLRVTGKPDAETLNV 86 Query: 199 VGKSRCGGKDVKPKNKRYNAANSKWDK-GTTYRFYIHDERHPTTMTLGEIQRALMIGLSY 375 + + RCG DV P S W+ TYR E + ++ ++ +A+ Sbjct: 87 MKQPRCGVPDVAPFV--LTPGKSCWENTNLTYRI----ENYTPDLSRADVDQAIEKAFQL 140 Query: 376 WSTAGDIYFAKVSNKQGSMFSLSFADAEHVQYDVHNNFVESRDPYPLDNVPGGVLAHAFF 555 WS + F KVS Q + +SF +H RD P D PGG LAHAF Sbjct: 141 WSNVTPLTFTKVSEGQADIM-ISFVRGDH------------RDNSPFDG-PGGNLAHAFQ 186 Query: 556 PR---NGNLHFDSAEEWTEGKEDGINLRLVAAHEMGHAIGLFHSNDPNALMAPWYGGYIS 726 P G+ HFD E WT +D NL VAAHE GH++GL HS D ALM Y Y Sbjct: 187 PGAGIGGDAHFDDDEWWTSNFQD-YNLYRVAAHEFGHSLGLAHSTDIGALM---YPSYTF 242 Query: 727 ENDFKLPDDDSQGAAFIYGTKNGKVFVRSNKKREQSANEKELCNITNINSAFQYSDDNIY 906 D +L DD G IYG + E ++ IT I + D Y Sbjct: 243 SGDVQLSQDDIDGIQAIYGPSQNPTQPVGPQTPEVCDSKLTFDAITTIRGEVMFFKDRFY 302 Query: 907 VVTDKNKVFRIIHSNNSFRVDASFKTLQLPNQLFDSDALFYDYSGRRFV-FFKGNRVFSA 1083 + T N ++ + N S QLPN L Y+ + R V FFKGN+ ++ Sbjct: 303 MRT--NPLYPEVELN-----FISVFWPQLPNGL----QAAYEVADRDEVRFFKGNKYWAV 351 Query: 1084 NGVKLENLTISELNVTRPGGFQGRV---TAAIVRDKTGKS 1194 G + L ++ R GF V AA+ + TGK+ Sbjct: 352 KGQDV--LRGYPRDIYRSFGFPRTVKSIDAAVSEEDTGKT 389
>sp|P45452|MMP13_HUMAN Collagenase 3 precursor (Matrix metalloproteinase-13) (MMP-13) Length = 471 Score = 128 bits (322), Expect = 4e-29 Identities = 114/395 (28%), Positives = 172/395 (43%), Gaps = 15/395 (3%) Frame = +1 Query: 55 DEKYVEKFLSDFYD-------YNENT-DRLNDAVKNIQKMAGLPTDGKINKELLAHVGKS 210 D ++ E++L +Y EN + + ++ +Q GL GK++ L + K Sbjct: 35 DLQFAERYLRSYYHPTNLAGILKENAASSMTERLREMQSFFGLEVTGKLDDNTLDVMKKP 94 Query: 211 RCGGKDVKPKNKRYNAANSKWDK-GTTYRFYIHDERHPTTMTLGEIQRALMIGLSYWSTA 387 RCG DV N KW K TYR + MT E+++A WS Sbjct: 95 RCGVPDVGEYN--VFPRTLKWSKMNLTYRIV----NYTPDMTHSEVEKAFKKAFKVWSDV 148 Query: 388 GDIYFAKVSNKQGSMFSLSFADAEHVQYDVHNNFVESRDPYPLDNVPGGVLAHAFFPR-- 561 + F ++ + + +SF EH + YP D P G+LAHAF P Sbjct: 149 TPLNFTRLHDGIADIM-ISFGIKEHGDF------------YPFDG-PSGLLAHAFPPGPN 194 Query: 562 -NGNLHFDSAEEWTEGKEDGINLRLVAAHEMGHAIGLFHSNDPNALMAPWYGGYISENDF 738 G+ HFD E WT + G NL LVAAHE GH++GL HS DP ALM P Y Y ++ F Sbjct: 195 YGGDAHFDDDETWTSSSK-GYNLFLVAAHEFGHSLGLDHSKDPGALMFPIY-TYTGKSHF 252 Query: 739 KLPDDDSQGAAFIYGTKNGKVFVRSNKKREQSANEKELCNITNINSAFQYSDDNIYVVTD 918 LPDDD QG +YG + + K ++ L IT++ D + Sbjct: 253 MLPDDDVQGIQSLYGPGDEDPNPKHPKTPDKCDPSLSLDAITSLRGETMIFKDRFFWRLH 312 Query: 919 KNKVFRIIHSNNSFRVDASFKTLQLPNQLFDSDALFYDYSGRRFVFFKGNRVFSANGVKL 1098 +V + SF +LPN++ DA + S F+G + ++ NG + Sbjct: 313 PQQVDAELFLTKSF-------WPELPNRI---DAAYEHPSHDLIFIFRGRKFWALNGYDI 362 Query: 1099 EN---LTISELNVTRPGGFQGRVTAAIVRDKTGKS 1194 ISEL + + +++AA+ + TGK+ Sbjct: 363 LEGYPKKISELGLPKE---VKKISAAVHFEDTGKT 394
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 174,492,514
Number of Sequences: 369166
Number of extensions: 3749745
Number of successful extensions: 10882
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9919
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10669
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 19315286550
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail