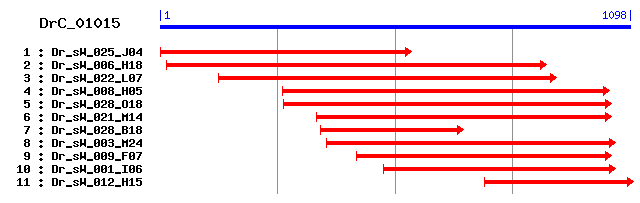
DrC_01015
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01015 (1098 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q19673|TYR3_CAEEL Putative tyrosinase-like protein tyr-3... 124 4e-28 sp|Q63722|JAG1_RAT Jagged-1 precursor (Jagged1) 72 4e-12 sp|Q9QXX0|JAG1_MOUSE Jagged-1 precursor (Jagged1) 71 5e-12 sp|Q19269|NAS14_CAEEL Zinc metalloproteinase nas-14 precurs... 69 2e-11 sp|Q90Y54|JAG1B_BRARE Jagged-1b precursor (Jagged1b) (Jagged3) 69 2e-11 sp|P78504|JAG1_HUMAN Jagged-1 precursor (Jagged1) (hJ1) (CD... 68 4e-11 sp|Q20191|NAS13_CAEEL Zinc metalloproteinase nas-13 precurs... 67 1e-10 sp|Q9QW30|NOTC2_RAT Neurogenic locus notch homolog protein ... 65 3e-10 sp|Q14162|SREC_HUMAN Endothelial cells scavenger receptor p... 64 8e-10 sp|Q9IAT6|DLLC_BRARE Delta-like protein C precursor (DeltaC... 63 1e-09
>sp|Q19673|TYR3_CAEEL Putative tyrosinase-like protein tyr-3 precursor Length = 683 Score = 124 bits (312), Expect = 4e-28 Identities = 66/200 (33%), Positives = 88/200 (44%), Gaps = 35/200 (17%) Frame = +1 Query: 268 CSDYNRNCTKWAREGLCEKNFNFMQKNCKFSCDACQDIGDGDNSNPNCLNSHESCDTWAD 447 C + N C W+ +G C+KN +M CK SC C N N C + H +C W+ Sbjct: 462 CFNENECCGPWSAKGECQKNPVYMNVWCKASCRQCTP---NYNINEECSDRHTNCAMWSR 518 Query: 448 RGECASNPGYMLVSCKKACNVCGKENK--------------------------------- 528 GEC NP +M +C+ +C CG+ Sbjct: 519 SGECNKNPLWMSENCRSSCQKCGRSRAATCGGGGGADSISNPTTMPPATNNGQQNTPCDS 578 Query: 529 --CADYRKRCPIMAKYGLCESDPRFMLQNCKESCGVCDGSSKNRPCVDANQDCAGWAASH 702 C + + CPI A+ G C S+P +M CK SCGVC + PC D + DCA WA Sbjct: 579 PMCYNEDQCCPIWAQRGQCRSNPGYMTCQCKVSCGVCRPNYVYGPCADYHYDCAAWARRG 638 Query: 703 ECETNPSWMLYNCKKSCNHC 762 EC N WM NC++SCN C Sbjct: 639 ECLKN-KWMPENCRRSCNTC 657
Score = 116 bits (290), Expect = 1e-25
Identities = 65/212 (30%), Positives = 88/212 (41%), Gaps = 36/212 (16%)
Frame = +1
Query: 406 NCLNSHESCDTWADRGECASNPGYMLVSCKKACNVCGKE----NKCADYRKRCPIMAKYG 573
+C N +E C W+ +GEC NP YM V CK +C C +C+D C + ++ G
Sbjct: 461 SCFNENECCGPWSAKGECQKNPVYMNVWCKASCRQCTPNYNINEECSDRHTNCAMWSRSG 520
Query: 574 LCESDPRFMLQNCKESCGVCDGS---------------------------SKNRPC---- 660
C +P +M +NC+ SC C S +N PC
Sbjct: 521 ECNKNPLWMSENCRSSCQKCGRSRAATCGGGGGADSISNPTTMPPATNNGQQNTPCDSPM 580
Query: 661 -VDANQDCAGWAASHECETNPSWMLYNCKKSCNHCRDARLESISNPNCKDLQIHCLYWAN 837
+ +Q C WA +C +NP +M CK SC CR + P C D C WA
Sbjct: 581 CYNEDQCCPIWAQRGQCRSNPGYMTCQCKVSCGVCRP---NYVYGP-CADYHYDCAAWAR 636
Query: 838 AGECQLNPGWAKQNCPFSCQHCGGCRDLIQNC 933
GEC N W +NC SC C + L C
Sbjct: 637 RGECLKNK-WMPENCRRSCNTCVNQQQLAARC 667
Score = 108 bits (269), Expect = 4e-23
Identities = 62/209 (29%), Positives = 85/209 (40%), Gaps = 41/209 (19%)
Frame = +1
Query: 31 NQYCQEWKKSGYCEKEPNTMKEYCSKTCGFCESETE-DPNCIDRDRNCQMWSDRGECGIN 207
N+ C W G C+K P M +C +C C + C DR NC MWS GEC N
Sbjct: 466 NECCGPWSAKGECQKNPVYMNVWCKASCRQCTPNYNINEECSDRHTNCAMWSRSGECNKN 525
Query: 208 PEAMLVTCKKTCNQCG--------------------------NPGK---------CSDYN 282
P M C+ +C +CG N G+ C + +
Sbjct: 526 PLWMSENCRSSCQKCGRSRAATCGGGGGADSISNPTTMPPATNNGQQNTPCDSPMCYNED 585
Query: 283 RNCTKWAREGLCEKNFNFMQKNCKFSCDACQDIGDGDNSNPN-----CLNSHESCDTWAD 447
+ C WA+ G C N +M CK SC C+ PN C + H C WA
Sbjct: 586 QCCPIWAQRGQCRSNPGYMTCQCKVSCGVCR---------PNYVYGPCADYHYDCAAWAR 636
Query: 448 RGECASNPGYMLVSCKKACNVCGKENKCA 534
RGEC N +M +C+++CN C + + A
Sbjct: 637 RGECLKNK-WMPENCRRSCNTCVNQQQLA 664
Score = 105 bits (263), Expect = 2e-22
Identities = 63/203 (31%), Positives = 89/203 (43%), Gaps = 35/203 (17%)
Frame = +1
Query: 511 CGKENKCADYRKRCPIMAKYGLCESDPRFMLQNCKESCGVCDGSSK-NRPCVDANQDCAG 687
C EN+C C + G C+ +P +M CK SC C + N C D + +CA
Sbjct: 462 CFNENEC------CGPWSAKGECQKNPVYMNVWCKASCRQCTPNYNINEECSDRHTNCAM 515
Query: 688 WAASHECETNPSWMLYNCKKSCNHCRDAR---------LESISNPN-------------- 798
W+ S EC NP WM NC+ SC C +R +SISNP
Sbjct: 516 WSRSGECNKNPLWMSENCRSSCQKCGRSRAATCGGGGGADSISNPTTMPPATNNGQQNTP 575
Query: 799 -----CKDLQIHCLYWANAGECQLNPGWAKQNCPFSCQHC------GGCRDLIQNCQNFA 945
C + C WA G+C+ NPG+ C SC C G C D +C +A
Sbjct: 576 CDSPMCYNEDQCCPIWAQRGQCRSNPGYMTCQCKVSCGVCRPNYVYGPCADYHYDCAAWA 635
Query: 946 NQGYCKTNEVYMNQSCKKTCGFC 1014
+G C N+ +M ++C+++C C
Sbjct: 636 RRGECLKNK-WMPENCRRSCNTC 657
Score = 88.6 bits (218), Expect = 3e-17
Identities = 48/155 (30%), Positives = 66/155 (42%), Gaps = 36/155 (23%)
Frame = +1
Query: 16 CEDKPNQYCQEWKKSGYCEKEPNTMKEYCSKTCGFC------------------------ 123
C D+ C W +SG C K P M E C +C C
Sbjct: 506 CSDRHTN-CAMWSRSGECNKNPLWMSENCRSSCQKCGRSRAATCGGGGGADSISNPTTMP 564
Query: 124 --------ESETEDPNCIDRDRNCQMWSDRGECGINPEAMLVTCKKTCNQCGNP----GK 267
+ + P C + D+ C +W+ RG+C NP M CK +C C P G
Sbjct: 565 PATNNGQQNTPCDSPMCYNEDQCCPIWAQRGQCRSNPGYMTCQCKVSCGVC-RPNYVYGP 623
Query: 268 CSDYNRNCTKWAREGLCEKNFNFMQKNCKFSCDAC 372
C+DY+ +C WAR G C KN +M +NC+ SC+ C
Sbjct: 624 CADYHYDCAAWARRGECLKN-KWMPENCRRSCNTC 657
>sp|Q63722|JAG1_RAT Jagged-1 precursor (Jagged1) Length = 1219 Score = 71.6 bits (174), Expect = 4e-12 Identities = 83/381 (21%), Positives = 131/381 (34%), Gaps = 47/381 (12%) Frame = +1 Query: 16 CEDKPNQYCQEWKKSGYCEKEPNTMKEYCSKTCG-----------------FCESETEDP 144 C+ N+ C E C K ++ CS G +C+ P Sbjct: 212 CDQNGNKTCMEGWMGPECNKA--ICRQGCSPKHGSCKLPGDCRCQYGWQGLYCDKCIPHP 269 Query: 145 NCIDRDRNCQMWSDRGECGINPEAMLVTCKKTCNQCGNPGKCSDYNRNCTKWAREGLCEK 324 C+ N + W C N L C K N CG C + + C Sbjct: 270 GCVHGTCN-EPWQCL--CETNWGGQL--CDKDLNYCGTHQPCLNRGTCSNTGPDKYQCSC 324 Query: 325 NFNFMQKNCKFSCDACQDIGDGDNSNPNCLNSHESCDTWADRGECASNPGYMLVSCKKAC 504 + NC+ + AC S+P ++ SC + EC +PG+ +C Sbjct: 325 PEGYSGPNCEIAEHACL-------SDP--CHNRGSCKETSSGFECECSPGWTGPTCSTNI 375 Query: 505 NVCGKENKCADYRKRCPIMAKYGLCESDPRFMLQNCKESCGVCDGSSKNRPCVDANQDCA 684 + C N + C + C P++ + C+ C+ +PCV+A + C Sbjct: 376 DDCSPNN--CSHGGTCQDLVNGFKCVCPPQWTGKTCQLDANECEA----KPCVNA-RSCK 428 Query: 685 GWAASHECETNPSWMLYNCKKSCNHC-----RDARLESISN------------PNC-KDL 810 AS+ C+ P WM NC + N C DA + N +C +D+ Sbjct: 429 NLIASYYCDCLPGWMGQNCDININDCLGQCQNDASCRDLVNGYRCICPPGYAGDHCERDI 488 Query: 811 QI----------HCLYWANAGECQLNPGWAKQNCPFSCQHC--GGCRDLIQNCQNFANQG 954 HC N +C G++ C +C C++ Q C N A+ Sbjct: 489 DECASNPCLNGGHCQNEINRFQCLCPTGFSGNLCQLDIDYCEPNPCQNGAQ-CYNRASDY 547 Query: 955 YCKTNEVYMNQSCKKTCGFCR 1017 +CK E Y ++C CR Sbjct: 548 FCKCPEDYEGKNCSHLKDHCR 568
Score = 61.6 bits (148), Expect = 4e-09
Identities = 81/366 (22%), Positives = 130/366 (35%), Gaps = 37/366 (10%)
Frame = +1
Query: 7 ASVCEDKP---NQYCQEWKKSGYCEKEPNTMKEYCSKTCGFCESETE-DPNCIDRDRNCQ 174
A+ CE KP + C+ S YC+ P M + C C + + D +C D +
Sbjct: 413 ANECEAKPCVNARSCKNLIASYYCDCLPGWMGQNCDININDCLGQCQNDASCRDLVNGYR 472
Query: 175 MWSDRGECGINPEAMLVTCKKTCNQCGNPGKC-SDYNRN---CTKWAREGLCEKNFNFMQ 342
G G + E + C N C N G C ++ NR C LC+ + ++ +
Sbjct: 473 CICPPGYAGDHCERDIDECAS--NPCLNGGHCQNEINRFQCLCPTGFSGNLCQLDIDYCE 530
Query: 343 KN-CKFSCDACQDIGDGDNSNPNCLNSHESCDTWADRGECASNPGYMLVSCKKAC----- 504
N C+ + Q + C +E + + C + P ++ SC A
Sbjct: 531 PNPCQ---NGAQCYNRASDYFCKCPEDYEGKNCSHLKDHCRTTPCEVIDSCTVAMASNDT 587
Query: 505 ---------NVCGKENKCADYRKRCPIMAKYGLCESDPRFMLQNCKESCGVCDGSSKNRP 657
NVCG KC + K+ C+ + F C E+ C+G+ P
Sbjct: 588 PEGVRYISSNVCGPHGKC-----KSESGGKF-TCDCNKGFTGTYCHENINDCEGN----P 637
Query: 658 CVDANQDCAGWAASHECETNPSWMLYNCKKSCNHCRDARLESISNP-----NCKDLQIHC 822
C + C S++C + W +C+ + N C NP C+DL
Sbjct: 638 CTNGGT-CIDGVNSYKCICSDGWEGAHCENNINDCS-------QNPCHYGGTCRDL---- 685
Query: 823 LYWANAGECQLNPGWAKQNC--------PFSCQHCGGCRDLIQNCQNFANQGYCKTN-EV 975
N C GW + C +C + G C D + + G+ T +
Sbjct: 686 ---VNDFYCDCKNGWKGKTCHSRDSQCDEATCNNGGTCYDEVDTFKCMCPGGWEGTTCNI 742
Query: 976 YMNQSC 993
N SC
Sbjct: 743 ARNSSC 748
Score = 50.8 bits (120), Expect = 7e-06
Identities = 55/260 (21%), Positives = 86/260 (33%), Gaps = 9/260 (3%)
Frame = +1
Query: 40 CQEWKKSGYCEKEPNTMKEYCSKTCGFCESETEDPNCIDRDRNCQMWSDRGECGINPEAM 219
C+E CE P CS C PN CQ + +C P+
Sbjct: 351 CKETSSGFECECSPGWTGPTCSTNIDDCS-----PNNCSHGGTCQDLVNGFKCVCPPQWT 405
Query: 220 LVTCKKTCNQCGNPGKCSDYNRNCTKWAREGLCEKNFNFMQKNCKFSCDACQDIGDGDNS 399
TC+ N+C K R+C C+ +M +NC + + C D S
Sbjct: 406 GKTCQLDANEC--EAKPCVNARSCKNLIASYYCDCLPGWMGQNCDININDCLGQCQNDAS 463
Query: 400 NPNCLNSHESCDTWADRGECASNPGYMLVSCKKACNVCGK---------ENKCADYRKRC 552
+ +N + C PGY C++ + C +N+ ++ C
Sbjct: 464 CRDLVNGYR----------CICPPGYAGDHCERDIDECASNPCLNGGHCQNEINRFQCLC 513
Query: 553 PIMAKYGLCESDPRFMLQNCKESCGVCDGSSKNRPCVDANQDCAGWAASHECETNPSWML 732
P LC+ D + N PC + Q C A+ + C+ +
Sbjct: 514 PTGFSGNLCQLDIDYCEPN---------------PCQNGAQ-CYNRASDYFCKCPEDYEG 557
Query: 733 YNCKKSCNHCRDARLESISN 792
NC +HCR E I +
Sbjct: 558 KNCSHLKDHCRTTPCEVIDS 577
>sp|Q9QXX0|JAG1_MOUSE Jagged-1 precursor (Jagged1) Length = 1218 Score = 71.2 bits (173), Expect = 5e-12 Identities = 83/381 (21%), Positives = 131/381 (34%), Gaps = 47/381 (12%) Frame = +1 Query: 16 CEDKPNQYCQEWKKSGYCEKEPNTMKEYCSKTCG-----------------FCESETEDP 144 C+ N+ C E C K ++ CS G +C+ P Sbjct: 212 CDQNGNKTCMEGWMGPDCNKA--ICRQGCSPKHGSCKLPGDCRCQYGWQGLYCDKCIPHP 269 Query: 145 NCIDRDRNCQMWSDRGECGINPEAMLVTCKKTCNQCGNPGKCSDYNRNCTKWAREGLCEK 324 C+ N + W C N L C K N CG C + + C Sbjct: 270 GCVHGTCN-EPWQCL--CETNWGGQL--CDKDLNYCGTHQPCLNRGTCSNTGPDKYQCSC 324 Query: 325 NFNFMQKNCKFSCDACQDIGDGDNSNPNCLNSHESCDTWADRGECASNPGYMLVSCKKAC 504 + NC+ + AC S+P ++ SC + EC +PG+ +C Sbjct: 325 PEGYSGPNCEIAEHACL-------SDP--CHNRGSCKETSSGFECECSPGWTGPTCSTNI 375 Query: 505 NVCGKENKCADYRKRCPIMAKYGLCESDPRFMLQNCKESCGVCDGSSKNRPCVDANQDCA 684 + C N + C + C P++ + C+ C+ +PCV+A + C Sbjct: 376 DDCSPNN--CSHGGTCQDLVNGFKCVCPPQWTGKTCQLDANECEA----KPCVNA-RSCK 428 Query: 685 GWAASHECETNPSWMLYNCKKSCNHC-----RDARLESISN------------PNC-KDL 810 AS+ C+ P WM NC + N C DA + N +C +D+ Sbjct: 429 NLIASYYCDCLPGWMGQNCDININDCLGQCQNDASCRDLVNGYRCICPPGYAGDHCERDI 488 Query: 811 QI----------HCLYWANAGECQLNPGWAKQNCPFSCQHC--GGCRDLIQNCQNFANQG 954 HC N +C G++ C +C C++ Q C N A+ Sbjct: 489 DECASNPCLNGGHCQNEINRFQCLCPTGFSGNLCQLDIDYCEPNPCQNGAQ-CYNRASDY 547 Query: 955 YCKTNEVYMNQSCKKTCGFCR 1017 +CK E Y ++C CR Sbjct: 548 FCKCPEDYEGKNCSHLKDHCR 568
Score = 56.2 bits (134), Expect = 2e-07
Identities = 84/367 (22%), Positives = 127/367 (34%), Gaps = 38/367 (10%)
Frame = +1
Query: 7 ASVCEDKP---NQYCQEWKKSGYCEKEPNTMKEYCSKTCGFCESETE-DPNCIDRDRNCQ 174
A+ CE KP + C+ S YC+ P M + C C + + D +C D +
Sbjct: 413 ANECEAKPCVNARSCKNLIASYYCDCLPGWMGQNCDININDCLGQCQNDASCRDLVNGYR 472
Query: 175 MWSDRGECGINPEAMLVTCKKTCNQCGNPGKC-SDYNRN---CTKWAREGLCEKNFNFMQ 342
G G + E + C N C N G C ++ NR C LC+ + ++ +
Sbjct: 473 CICPPGYAGDHCERDIDECAS--NPCLNGGHCQNEINRFQCLCPTGFSGNLCQLDIDYCE 530
Query: 343 KN-CKFSCDACQDIGD------GDNSNPNCLNSHESCDTWADR--GEC----ASNP---G 474
N C+ D D NC + + C T C ASN G
Sbjct: 531 PNPCQNGAQCYNRASDYFCKCPEDYEGKNCSHLKDHCRTTTCEVIDSCTVAMASNDTPEG 590
Query: 475 YMLVSCKKACNVCGKENKCADYRKRCPIMAKYGLCESDPRFMLQNCKESCGVCDGSSKNR 654
+S NVCG KC K+ C+ + F C E+ C+ +
Sbjct: 591 VRYISS----NVCGPHGKCKSQSG-----GKF-TCDCNKGFTGTYCHENINDCESN---- 636
Query: 655 PCVDANQDCAGWAASHECETNPSWMLYNCKKSCNHCRDARLESISNP-----NCKDLQIH 819
PC + C S++C + W +C+ + N C NP C+DL
Sbjct: 637 PCKNGGT-CIDGVNSYKCICSDGWEGAHCENNINDCS-------QNPCHYGGTCRDL--- 685
Query: 820 CLYWANAGECQLNPGWAKQNC--------PFSCQHCGGCRDLIQNCQNFANQGYCKTN-E 972
N C GW + C +C + G C D + + G+ T
Sbjct: 686 ----VNDFYCDCKNGWKGKTCHSRDSQCDEATCNNGGTCYDEVDTFKCMCPGGWEGTTCN 741
Query: 973 VYMNQSC 993
+ N SC
Sbjct: 742 IARNSSC 748
Score = 51.2 bits (121), Expect = 5e-06
Identities = 55/260 (21%), Positives = 86/260 (33%), Gaps = 9/260 (3%)
Frame = +1
Query: 40 CQEWKKSGYCEKEPNTMKEYCSKTCGFCESETEDPNCIDRDRNCQMWSDRGECGINPEAM 219
C+E CE P CS C PN CQ + +C P+
Sbjct: 351 CKETSSGFECECSPGWTGPTCSTNIDDCS-----PNNCSHGGTCQDLVNGFKCVCPPQWT 405
Query: 220 LVTCKKTCNQCGNPGKCSDYNRNCTKWAREGLCEKNFNFMQKNCKFSCDACQDIGDGDNS 399
TC+ N+C K R+C C+ +M +NC + + C D S
Sbjct: 406 GKTCQLDANEC--EAKPCVNARSCKNLIASYYCDCLPGWMGQNCDININDCLGQCQNDAS 463
Query: 400 NPNCLNSHESCDTWADRGECASNPGYMLVSCKKACNVCGK---------ENKCADYRKRC 552
+ +N + C PGY C++ + C +N+ ++ C
Sbjct: 464 CRDLVNGYR----------CICPPGYAGDHCERDIDECASNPCLNGGHCQNEINRFQCLC 513
Query: 553 PIMAKYGLCESDPRFMLQNCKESCGVCDGSSKNRPCVDANQDCAGWAASHECETNPSWML 732
P LC+ D + N PC + Q C A+ + C+ +
Sbjct: 514 PTGFSGNLCQLDIDYCEPN---------------PCQNGAQ-CYNRASDYFCKCPEDYEG 557
Query: 733 YNCKKSCNHCRDARLESISN 792
NC +HCR E I +
Sbjct: 558 KNCSHLKDHCRTTTCEVIDS 577
>sp|Q19269|NAS14_CAEEL Zinc metalloproteinase nas-14 precursor (Nematode astacin 14) Length = 503 Score = 69.3 bits (168), Expect = 2e-11 Identities = 41/125 (32%), Positives = 53/125 (42%), Gaps = 6/125 (4%) Frame = +1 Query: 16 CEDKPNQYCQEWKKSGYCEKEPNTMKEYCSKTCGFCESE-----TEDPNCIDRDRNCQMW 180 CED N +C W++ G+C+ M YC K C CE E T P + R++ + Sbjct: 380 CEDL-NAHCGMWEQLGHCQHSVKYMAHYCRKACNLCEVEVTTTTTTTPKPVPRNKEKENK 438 Query: 181 SDRGECGINPEAMLVTCKKTCNQCGNPG-KCSDYNRNCTKWAREGLCEKNFNFMQKNCKF 357 S A T K T P KC D N C+ WA+ G C FM+ CK Sbjct: 439 SASSTTRGTSTATSTTPKTTTTTTSAPKEKCEDKNLFCSYWAKIGECNSESKFMKIFCKA 498 Query: 358 SCDAC 372 SC C Sbjct: 499 SCGKC 503
Score = 57.8 bits (138), Expect = 5e-08
Identities = 36/144 (25%), Positives = 57/144 (39%), Gaps = 2/144 (1%)
Frame = +1
Query: 208 PEAMLVTCKKTCNQCGNPGKCSDYNRNCTKWAREGLCEKNFNFMQKNCKFSCDACQ-DIG 384
P T K + N KC D N +C W + G C+ + +M C+ +C+ C+ ++
Sbjct: 361 PTTTTTTTPKPVERSRNK-KCEDLNAHCGMWEQLGHCQHSVKYMAHYCRKACNLCEVEVT 419
Query: 385 DGDNSNPNCL-NSHESCDTWADRGECASNPGYMLVSCKKACNVCGKENKCADYRKRCPIM 561
+ P + + E + A ++ + KC D C
Sbjct: 420 TTTTTTPKPVPRNKEKENKSASSTTRGTSTATSTTPKTTTTTTSAPKEKCEDKNLFCSYW 479
Query: 562 AKYGLCESDPRFMLQNCKESCGVC 633
AK G C S+ +FM CK SCG C
Sbjct: 480 AKIGECNSESKFMKIFCKASCGKC 503
Score = 52.8 bits (125), Expect = 2e-06
Identities = 35/137 (25%), Positives = 47/137 (34%), Gaps = 16/137 (11%)
Frame = +1
Query: 400 NPNCLNSHESCDTWADRGECASNPGYMLVSCKKACNVCGKENKCADYRKRCPIMAKYGLC 579
N C + + C W G C + YM C+KACN+C E P+
Sbjct: 377 NKKCEDLNAHCGMWEQLGHCQHSVKYMAHYCRKACNLCEVEVTTTTTTTPKPV------- 429
Query: 580 ESDPRFMLQNCKESCGVCDGSSK----------------NRPCVDANQDCAGWAASHECE 711
PR + K + G+S C D N C+ WA EC
Sbjct: 430 ---PRNKEKENKSASSTTRGTSTATSTTPKTTTTTTSAPKEKCEDKNLFCSYWAKIGECN 486
Query: 712 TNPSWMLYNCKKSCNHC 762
+ +M CK SC C
Sbjct: 487 SESKFMKIFCKASCGKC 503
Score = 50.4 bits (119), Expect = 9e-06
Identities = 31/129 (24%), Positives = 43/129 (33%), Gaps = 42/129 (32%)
Frame = +1
Query: 643 SKNRPCVDANQDCAGWAASHECETNPSWMLYNCKKSCNHC----------------RDAR 774
S+N+ C D N C W C+ + +M + C+K+CN C R+
Sbjct: 375 SRNKKCEDLNAHCGMWEQLGHCQHSVKYMAHYCRKACNLCEVEVTTTTTTTPKPVPRNKE 434
Query: 775 LE--------------------------SISNPNCKDLQIHCLYWANAGECQLNPGWAKQ 876
E S C+D + C YWA GEC + K
Sbjct: 435 KENKSASSTTRGTSTATSTTPKTTTTTTSAPKEKCEDKNLFCSYWAKIGECNSESKFMKI 494
Query: 877 NCPFSCQHC 903
C SC C
Sbjct: 495 FCKASCGKC 503
Score = 46.6 bits (109), Expect = 1e-04
Identities = 31/139 (22%), Positives = 50/139 (35%), Gaps = 12/139 (8%)
Frame = +1
Query: 133 TEDPNCIDRDRNCQMWSDRGECGINPEAMLVTCKKTCNQCGNPGKCSDYNRNCTKWAREG 312
+ + C D + +C MW G C + + M C+K CN C + T
Sbjct: 375 SRNKKCEDLNAHCGMWEQLGHCQHSVKYMAHYCRKACNLC-------EVEVTTTTTTTPK 427
Query: 313 LCEKNFNFMQKNCKFSCDACQDIGDGDNSNP------------NCLNSHESCDTWADRGE 456
+N +K K + + ++ P C + + C WA GE
Sbjct: 428 PVPRN---KEKENKSASSTTRGTSTATSTTPKTTTTTTSAPKEKCEDKNLFCSYWAKIGE 484
Query: 457 CASNPGYMLVSCKKACNVC 513
C S +M + CK +C C
Sbjct: 485 CNSESKFMKIFCKASCGKC 503
Score = 35.0 bits (79), Expect = 0.38
Identities = 14/43 (32%), Positives = 19/43 (44%)
Frame = +1
Query: 775 LESISNPNCKDLQIHCLYWANAGECQLNPGWAKQNCPFSCQHC 903
+E N C+DL HC W G CQ + + C +C C
Sbjct: 372 VERSRNKKCEDLNAHCGMWEQLGHCQHSVKYMAHYCRKACNLC 414
Score = 33.1 bits (74), Expect = 1.4
Identities = 12/35 (34%), Positives = 18/35 (51%)
Frame = +1
Query: 910 CRDLIQNCQNFANQGYCKTNEVYMNQSCKKTCGFC 1014
C DL +C + G+C+ + YM C+K C C
Sbjct: 380 CEDLNAHCGMWEQLGHCQHSVKYMAHYCRKACNLC 414
>sp|Q90Y54|JAG1B_BRARE Jagged-1b precursor (Jagged1b) (Jagged3) Length = 1213 Score = 68.9 bits (167), Expect = 2e-11 Identities = 69/324 (21%), Positives = 113/324 (34%), Gaps = 7/324 (2%) Frame = +1 Query: 67 CEKEPNTMKEYCSKTCGFCESETEDPNCIDRDRNCQMWSDRGECGINPEAMLVTCKKT-- 240 C + N + C K +C T P C++ D+ +C V C++ Sbjct: 279 CLCDTNWGGQLCDKDLNYCG--THQP-CLNGGTCSNTGPDKYQCSCEDGYSGVNCERAEH 335 Query: 241 ---CNQCGNPGKCSDYNRNCTKWAREGLCEKNFNFMQKNCKFSCDACQDIGDGDNSNPNC 411 N C N G C + ++ C + +C+ + D C PN Sbjct: 336 ACLSNPCANGGTCKETSQGYE-------CHCAIGWSGTSCEINVDDC---------TPNQ 379 Query: 412 LNSHESCDTWADRGECASNPGYMLVSCKKACNVCGKENKCADYRKRCPIMAKYGLCESDP 591 +C + +CA P + +C+ N C E+K K C + CE P Sbjct: 380 CKHGGTCQDLVNGFKCACPPHWTGKTCQIDANEC--EDKPCVNAKSCHNLIGAYFCECLP 437 Query: 592 RFMLQNCKESCGVCDGSSKNRPCVDANQDCAGWAASHECETNPSWMLYNCKKSCNHCRDA 771 + QNC + C G N C + C P + C+K + C A Sbjct: 438 GWSGQNCDININDCKGQCLN------GGTCKDLVNGYRCLCPPGYTGEQCEKDVDEC--A 489 Query: 772 RLESISNPNCKDLQIHCLYWANAGECQLNPGWAKQNCPFSCQHC--GGCRDLIQNCQNFA 945 ++ C+D N +C G++ Q C +C C++ Q C N A Sbjct: 490 SSPCLNGGRCQDE-------VNGFQCLCPAGFSGQLCQLDIDYCKPNPCQNGAQ-CFNLA 541 Query: 946 NQGYCKTNEVYMNQSCKKTCGFCR 1017 + +CK + Y ++C CR Sbjct: 542 SDYFCKCPDDYEGKNCSHLKDHCR 565
Score = 49.3 bits (116), Expect = 2e-05
Identities = 51/237 (21%), Positives = 81/237 (34%), Gaps = 2/237 (0%)
Frame = +1
Query: 67 CEKEPNTMKEYCSKTCGFCESET--EDPNCIDRDRNCQMWSDRGECGINPEAMLVTCKKT 240
CE + YC + CES CID+ Q G G++ E + C +
Sbjct: 612 CECQEGFRGTYCHENINDCESNPCRNGGTCIDKVNVYQCICADGWEGVHCEINIDDC--S 669
Query: 241 CNQCGNPGKCSDYNRNCTKWAREGLCEKNFNFMQKNCKFSCDACQDIGDGDNSNPNCLNS 420
N C N G C D + CE + K C C + N+
Sbjct: 670 LNPCLNKGACQDL-------VNDFYCECRNGWKGKTCHSRDSQCDEA---------TCNN 713
Query: 421 HESCDTWADRGECASNPGYMLVSCKKACNVCGKENKCADYRKRCPIMAKYGLCESDPRFM 600
+C D +C +PG+ +C A N N C + C + C +
Sbjct: 714 GGTCHDEGDTFKCRCSPGWEGATCNIAKNSSCLPNPC-ENGGTCVVNGDSFNCVCKEGWE 772
Query: 601 LQNCKESCGVCDGSSKNRPCVDANQDCAGWAASHECETNPSWMLYNCKKSCNHCRDA 771
C E+ C+ PC ++ C + CE P + +C+ + N C+ +
Sbjct: 773 GSTCTENTNDCN----PHPCYNSG-TCVDGENWYRCECAPGFAGPDCRININECQSS 824
Score = 48.9 bits (115), Expect = 3e-05
Identities = 83/365 (22%), Positives = 124/365 (33%), Gaps = 36/365 (9%)
Frame = +1
Query: 28 PNQYCQEWKKSGYCEKEPNTMKEYC-----SKTCGFCESETEDPNCIDRDRNCQMWSDRG 192
PNQ K G C+ N K C KTC +E ED C++ ++C
Sbjct: 377 PNQC----KHGGTCQDLVNGFKCACPPHWTGKTCQIDANECEDKPCVNA-KSCHNLIGAY 431
Query: 193 EC-------GINPEAMLVTCKKTCNQCGNPGKCSDYNRNCTKWAREGLCEKNFNFMQKNC 351
C G N + + CK QC N G C D LC + Q C
Sbjct: 432 FCECLPGWSGQNCDININDCK---GQCLNGGTCKDLVNGY-----RCLCPPGYTGEQ--C 481
Query: 352 KFSCDACQDIGDGDNSNPNCLNSHESCDTWADRGECASNPGYMLVSCKKACNVCGKENKC 531
+ D C S+P CLN C + +C G+ C+ + C K N C
Sbjct: 482 EKDVDECA-------SSP-CLNGGR-CQDEVNGFQCLCPAGFSGQLCQLDIDYC-KPNPC 531
Query: 532 ADYRKRCPIMAKYGLCESDPRFMLQNCKE--------SCGVCDG-----SSKNRP----- 657
+ +C +A C+ + +NC SC V D +S + P
Sbjct: 532 QN-GAQCFNLASDYFCKCPDDYEGKNCSHLKDHCRTTSCQVIDSCTVAVASNSTPEGVRY 590
Query: 658 -----CVDANQDCAGWAASHECETNPSWMLYNCKKSCNHCRDARLESISNPNCKDLQIHC 822
C + + CE + C ++ N C SNP C++ C
Sbjct: 591 ISSNVCGPHGRCRSQAGGQFTCECQEGFRGTYCHENINDCE-------SNP-CRNGGT-C 641
Query: 823 LYWANAGECQLNPGWAKQNCPFSCQHCGGCRDLIQN-CQNFANQGYCKTNEVYMNQSCKK 999
+ N +C GW +C + C L + CQ+ N YC+ + ++C
Sbjct: 642 IDKVNVYQCICADGWEGVHCEINIDDCSLNPCLNKGACQDLVNDFYCECRNGWKGKTCHS 701
Query: 1000 TCGFC 1014
C
Sbjct: 702 RDSQC 706
>sp|P78504|JAG1_HUMAN Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen) Length = 1218 Score = 68.2 bits (165), Expect = 4e-11 Identities = 86/386 (22%), Positives = 138/386 (35%), Gaps = 52/386 (13%) Frame = +1 Query: 16 CEDKPNQYCQEWKKSGYCEKEPNTMKEYCSKTCGFCESETEDPNCIDRDRNCQMWSDRGE 195 C+ N+ C E C + ++ CS G C+ + D CQ G Sbjct: 212 CDQNGNKTCMEGWMGPECNRA--ICRQGCSPKHGSCK--------LPGDCRCQY----GW 257 Query: 196 CGINPEAMLVTCKKTCNQCGNPGKCSDYNRNC-TKWAREGLCEKNFNFM--QKNCKFSCD 366 G+ + + C P +C C T W + LC+K+ N+ + C + Sbjct: 258 QGLYCDKCIPHPGCVHGICNEPWQCL-----CETNWGGQ-LCDKDLNYCGTHQPC-LNGG 310 Query: 367 ACQDIGDGDN--------SNPNCLNSHESC--DTWADRG---------ECASNPGYMLVS 489 C + G S PNC + +C D +RG EC +PG+ + Sbjct: 311 TCSNTGPDKYQCSCPEGYSGPNCEIAEHACLSDPCHNRGSCKETSLGFECECSPGWTGPT 370 Query: 490 CKKACNVCGKENKCADYRKRCPIMAKYGLCESDPRFMLQNCKESCGVCDGSSKNRPCVDA 669 C + C N + C + C P++ + C+ C+ +PCV+A Sbjct: 371 CSTNIDDCSPNN--CSHGGTCQDLVNGFKCVCPPQWTGKTCQLDANECEA----KPCVNA 424 Query: 670 NQDCAGWAASHECETNPSWMLYNCKKSCNHC-----RDARLESISN------------PN 798 + C AS+ C+ P WM NC + N C DA + N + Sbjct: 425 -KSCKNLIASYYCDCLPGWMGQNCDININDCLGQCQNDASCRDLVNGYRCICPPGYAGDH 483 Query: 799 C-KDLQI----------HCLYWANAGECQLNPGWAKQNCPFSCQHC--GGCRDLIQNCQN 939 C +D+ HC N +C G++ C +C C++ Q C N Sbjct: 484 CERDIDECASNPCLNGGHCQNEINRFQCLCPTGFSGNLCQLDIDYCEPNPCQNGAQ-CYN 542 Query: 940 FANQGYCKTNEVYMNQSCKKTCGFCR 1017 A+ +CK E Y ++C CR Sbjct: 543 RASDYFCKCPEDYEGKNCSHLKDHCR 568
Score = 50.8 bits (120), Expect = 7e-06
Identities = 75/366 (20%), Positives = 122/366 (33%), Gaps = 40/366 (10%)
Frame = +1
Query: 40 CQEWKKSGYCEKEPNTMKEYCSKTCGFCESETEDPNCIDRDRNCQMWSDRGECGINPEAM 219
C+E CE P CS C PN CQ + +C P+
Sbjct: 351 CKETSLGFECECSPGWTGPTCSTNIDDCS-----PNNCSHGGTCQDLVNGFKCVCPPQWT 405
Query: 220 LVTCKKTCNQCGNPGKCSDYNRNCTKWAREGLCEKNFNFMQKNCKFSCDACQDIGDGDNS 399
TC+ N+C K ++C C+ +M +NC + + C D S
Sbjct: 406 GKTCQLDANEC--EAKPCVNAKSCKNLIASYYCDCLPGWMGQNCDININDCLGQCQNDAS 463
Query: 400 NPNCLNSHESCDTWADRGECASNPGYMLVSCKKACNVCGK---------ENKCADYRKRC 552
+ +N + C PGY C++ + C +N+ ++ C
Sbjct: 464 CRDLVNGYR----------CICPPGYAGDHCERDIDECASNPCLNGGHCQNEINRFQCLC 513
Query: 553 PIMAKYGLCESDPRFMLQNCKESCGVCDGSSKNRPCVDANQDCAGWAASHECETNPSWML 732
P LC+ D + N PC + Q C A+ + C+ +
Sbjct: 514 PTGFSGNLCQLDIDYCEPN---------------PCQNGAQ-CYNRASDYFCKCPEDYEG 557
Query: 733 YNCKKSCNHCRDARLESI--------SNPNCKDLQI--------HCLYWANAG---ECQL 855
NC +HCR E I SN + ++ H + +G C
Sbjct: 558 KNCSHLKDHCRTTPCEVIDSCTVAMASNDTPEGVRYISSNVCGPHGKCKSQSGGKFTCDC 617
Query: 856 NPGWAKQNCPFS--------CQHCGGCRDLIQNCQNFANQG----YCKTNEVYMNQSCKK 999
N G+ C + C++ G C D + + + + G YC+TN +Q+
Sbjct: 618 NKGFTGTYCHENINDCESNPCRNGGTCIDGVNSYKCICSDGWEGAYCETNINDCSQNPCH 677
Query: 1000 TCGFCR 1017
G CR
Sbjct: 678 NGGTCR 683
>sp|Q20191|NAS13_CAEEL Zinc metalloproteinase nas-13 precursor (Nematode astacin 13) Length = 527 Score = 66.6 bits (161), Expect = 1e-10 Identities = 36/84 (42%), Positives = 41/84 (48%), Gaps = 5/84 (5%) Frame = +1 Query: 526 KCADYRKRCPIMAKYGLCES--DPRFMLQNCKESCGVCDGSSKNRP-CVDANQDCAGWAA 696 KC D RK C +A+ G CES RFM +NC SCG C K + C DA C WA Sbjct: 444 KCEDRRKDCEFLARAGHCESRFSIRFMTENCANSCGKCIAEEKRKEVCEDARTWCERWAN 503 Query: 697 SHECETN--PSWMLYNCKKSCNHC 762 S C +M C KSCN C Sbjct: 504 SGMCNQTVFKDYMRQKCAKSCNFC 527
Score = 58.2 bits (139), Expect = 4e-08
Identities = 32/88 (36%), Positives = 43/88 (48%), Gaps = 4/88 (4%)
Frame = +1
Query: 262 GKCSDYNRNCTKWAREGLCEKNFN--FMQKNCKFSCDACQDIGDGDNSNPNCLNSHESCD 435
GKC D ++C AR G CE F+ FM +NC SC C I + + C ++ C+
Sbjct: 443 GKCEDRRKDCEFLARAGHCESRFSIRFMTENCANSCGKC--IAE-EKRKEVCEDARTWCE 499
Query: 436 TWADRGECASN--PGYMLVSCKKACNVC 513
WA+ G C YM C K+CN C
Sbjct: 500 RWANSGMCNQTVFKDYMRQKCAKSCNFC 527
Score = 49.3 bits (116), Expect = 2e-05
Identities = 27/83 (32%), Positives = 38/83 (45%), Gaps = 11/83 (13%)
Frame = +1
Query: 799 CKDLQIHCLYWANAGECQ--LNPGWAKQNCPFSCQHCGG-------CRDLIQNCQNFANQ 951
C+D + C + A AG C+ + + +NC SC C C D C+ +AN
Sbjct: 445 CEDRRKDCEFLARAGHCESRFSIRFMTENCANSCGKCIAEEKRKEVCEDARTWCERWANS 504
Query: 952 GYCKTN--EVYMNQSCKKTCGFC 1014
G C + YM Q C K+C FC
Sbjct: 505 GMCNQTVFKDYMRQKCAKSCNFC 527
Score = 47.8 bits (112), Expect = 6e-05
Identities = 28/86 (32%), Positives = 38/86 (44%), Gaps = 4/86 (4%)
Frame = +1
Query: 658 CVDANQDCAGWAASHECETNPS--WMLYNCKKSCNHCRDARLESISNPNCKDLQIHCLYW 831
C D +DC A + CE+ S +M NC SC C E C+D + C W
Sbjct: 445 CEDRRKDCEFLARAGHCESRFSIRFMTENCANSCGKCI---AEEKRKEVCEDARTWCERW 501
Query: 832 ANAGECQ--LNPGWAKQNCPFSCQHC 903
AN+G C + + +Q C SC C
Sbjct: 502 ANSGMCNQTVFKDYMRQKCAKSCNFC 527
Score = 46.2 bits (108), Expect = 2e-04
Identities = 26/84 (30%), Positives = 42/84 (50%), Gaps = 5/84 (5%)
Frame = +1
Query: 16 CEDKPNQYCQEWKKSGYCEKEPNT--MKEYCSKTCGFCESETEDPN-CIDRDRNCQMWSD 186
CED+ C+ ++G+CE + M E C+ +CG C +E + C D C+ W++
Sbjct: 445 CEDRRKD-CEFLARAGHCESRFSIRFMTENCANSCGKCIAEEKRKEVCEDARTWCERWAN 503
Query: 187 RGECG--INPEAMLVTCKKTCNQC 252
G C + + M C K+CN C
Sbjct: 504 SGMCNQTVFKDYMRQKCAKSCNFC 527
Score = 42.0 bits (97), Expect = 0.003
Identities = 25/83 (30%), Positives = 34/83 (40%), Gaps = 8/83 (9%)
Frame = +1
Query: 409 CLNSHESCDTWADRGECASNPG--YMLVSCKKACNVCGKENK----CADYRKRCPIMAKY 570
C + + C+ A G C S +M +C +C C E K C D R C A
Sbjct: 445 CEDRRKDCEFLARAGHCESRFSIRFMTENCANSCGKCIAEEKRKEVCEDARTWCERWANS 504
Query: 571 GLCESD--PRFMLQNCKESCGVC 633
G+C +M Q C +SC C
Sbjct: 505 GMCNQTVFKDYMRQKCAKSCNFC 527
Score = 36.6 bits (83), Expect = 0.13
Identities = 15/39 (38%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Frame = +1
Query: 13 VCEDKPNQYCQEWKKSGYCEKE--PNTMKEYCSKTCGFC 123
VCED +C+ W SG C + + M++ C+K+C FC
Sbjct: 490 VCEDA-RTWCERWANSGMCNQTVFKDYMRQKCAKSCNFC 527
Score = 31.2 bits (69), Expect = 5.5
Identities = 12/39 (30%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Frame = +1
Query: 904 GGCRDLIQNCQNFANQGYCKT--NEVYMNQSCKKTCGFC 1014
G C D ++C+ A G+C++ + +M ++C +CG C
Sbjct: 443 GKCEDRRKDCEFLARAGHCESRFSIRFMTENCANSCGKC 481
>sp|Q9QW30|NOTC2_RAT Neurogenic locus notch homolog protein 2 precursor (Notch 2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain] Length = 2471 Score = 65.1 bits (157), Expect = 3e-10 Identities = 83/339 (24%), Positives = 121/339 (35%), Gaps = 23/339 (6%) Frame = +1 Query: 67 CEKEPNTMKEYCSK--TCGFCESETE--DPNCIDRDRNCQMWSDRGECGINPEAMLVTCK 234 C PN + C+ T C+ + DP+ + CQ D C NP M C Sbjct: 547 CIDHPNGYECQCATGFTGTLCDENIDNCDPDPCHHGQ-CQDGIDSYTCICNPGYMGAICS 605 Query: 235 KTCNQCG-----NPGKCSD----YNRNCTKWAREGLCEKNFNFMQKNCKFSCDACQDIGD 387 ++C N G+C D Y NC GL NC+ + D C Sbjct: 606 DQIDECYSSPCLNDGRCIDLVNGYQCNCQP-GTSGL----------NCEINFDDCA---- 650 Query: 388 GDNSNPNCLNSHESCDTWADRGECASNPGYMLVSC-----KKACNVCGKENKCAD----Y 540 SNP CL H +C +R C +PG+ C + A N C K+ C + + Sbjct: 651 ---SNP-CL--HGACVDGINRYSCVCSPGFTGQRCNIDIDECASNPCRKDATCINDVNGF 704 Query: 541 RKRCPIMAKYGLCESDPRFMLQNCKESCGVCDGSSKNRPCVDANQDCAGWAASHECETNP 720 R CP + C S + C S PC+ N C G + ++C + Sbjct: 705 RCMCPEGPHHPSCYSQ----VNECLSS-----------PCIHGN--CTGGLSGYKCLCDA 747 Query: 721 SWMLYNCKKSCNHCRDARLESISNPNCKDLQIHCLYWANAGECQLNPGWAKQNCPFSCQH 900 W+ NC+ N C +SNP C++ C N C G+ NC + Sbjct: 748 GWVGINCEVDKNEC-------LSNP-CQNGGT-CNNLVNGYRCTCKKGFKGYNCQVNIDE 798 Query: 901 CGGCRDLIQ-NCQNFANQGYCKTNEVYMNQSCKKTCGFC 1014 C L Q C + + C Y ++C+ C Sbjct: 799 CASNPCLNQGTCLDDVSGYTCHCMLPYTGKNCQTVLAPC 837
Score = 54.3 bits (129), Expect = 6e-07
Identities = 85/360 (23%), Positives = 113/360 (31%), Gaps = 33/360 (9%)
Frame = +1
Query: 16 CEDKP---NQYCQEWKKSGYCEKEPNTMKEYCSKTCGFCESET--EDPNCIDRDRNCQMW 180
C KP N C + S CE P C + C + +C+D+
Sbjct: 877 CVSKPCMNNGICHNTQGSYMCECPPGFSGMDCEEDINDCLANPCQNGGSCVDKVNTFSCL 936
Query: 181 SDRGECGINPEAMLVTCKKTCNQCGNPGKCSDYNRNCTKWAREGL----CEKNFNFMQKN 348
G G + + C C N G CSDY + T G CE N + ++
Sbjct: 937 CLPGFVGDKCQTDMNECLS--EPCKNGGTCSDYVNSYTCTCPAGFHGVHCENNIDECTES 994
Query: 349 CKFSCDACQDIGDGDNS----------NPNCLNSHESCDTWADRGECASNPGYMLVSCKK 498
F+ C D G NS P CL+ D EC+SNP
Sbjct: 995 SCFNGGTCVD---GINSFSCLCPVGFTGPFCLH---------DINECSSNP--------- 1033
Query: 499 ACNVCGKENKCAD----YRKRCPIMAKYGLCESDPRFMLQNCKESCGVCDGSSKNRPCVD 666
C C D YR CP+ + +NC+ +C S PC +
Sbjct: 1034 ----CLNSGTCVDGLGTYRCTCPL-----------GYTGKNCQTLVNLCSPS----PCKN 1074
Query: 667 ANQDCAGWAASHECETNPSWMLYNCKKSCNHCRDARLESISNPN--CKDLQIHCLYWANA 840
CA A C P W C C+ A L+ C+ I C+ N
Sbjct: 1075 KGT-CAQEKARPRCLCPPGWDGAYCDVLNVSCKAAALQKGVPVEHLCQHSGI-CINAGNT 1132
Query: 841 GECQLNPGWAKQNCPFS--------CQHCGGCRDLIQNCQNFANQGYCKTNEVYMNQSCK 996
CQ G+ C CQH C D I + GY N Y C+
Sbjct: 1133 HHCQCPLGYTGSYCEEQLDECASNPCQHGATCSDFIGGYRCECVPGYQGVNCEYEVDECQ 1192
Score = 51.2 bits (121), Expect = 5e-06
Identities = 67/308 (21%), Positives = 103/308 (33%), Gaps = 10/308 (3%)
Frame = +1
Query: 100 CSKTCGFCESETEDP-----NCIDRDRNCQMWSDRGECGINPEAMLVTCKKTCNQCGNPG 264
C++ C +P C++ D +G G E + C + C N
Sbjct: 412 CTEDVDECAMANSNPCEHAGKCVNTDGAFHCECLKGYAGPRCEMDINECHS--DPCQNDA 469
Query: 265 KCSDYNRNCTKWAREGLCEKNFNFMQKNCKFSCDACQDIGDGDNSNPNCLNSHESCDTWA 444
C D T G F +C+ + CQ SNP C+N+ + D
Sbjct: 470 TCLDKIGGFTCLCMPG-------FKGVHCELEVNECQ-------SNP-CVNNGQCVDK-V 513
Query: 445 DRGECASNPGYMLVSCKKACNVCGKEN-----KCADYRKRCPIMAKYGLCESDPRFMLQN 609
+R +C PG+ C+ + C KC D+ C+ F
Sbjct: 514 NRFQCLCPPGFTGPVCQIDIDDCSSTPCLNGAKCIDHPNGYE-------CQCATGFTGTL 566
Query: 610 CKESCGVCDGSSKNRPCVDANQDCAGWAASHECETNPSWMLYNCKKSCNHCRDARLESIS 789
C E+ CD PC + C S+ C NP +M C + C S
Sbjct: 567 CDENIDNCDPD----PC--HHGQCQDGIDSYTCICNPGYMGAICSDQIDEC-------YS 613
Query: 790 NPNCKDLQIHCLYWANAGECQLNPGWAKQNCPFSCQHCGGCRDLIQNCQNFANQGYCKTN 969
+P D + C+ N +C PG + NC + C L C + N+ C +
Sbjct: 614 SPCLNDGR--CIDLVNGYQCNCQPGTSGLNCEINFDDCASNPCLHGACVDGINRYSCVCS 671
Query: 970 EVYMNQSC 993
+ Q C
Sbjct: 672 PGFTGQRC 679
Score = 50.8 bits (120), Expect = 7e-06
Identities = 77/342 (22%), Positives = 114/342 (33%), Gaps = 23/342 (6%)
Frame = +1
Query: 58 SGYCEKEPNTMKEYCSKTCGFCESETEDPNCIDRDRNC-----------------QMWSD 186
S C K+ + + C C P+C + C + D
Sbjct: 688 SNPCRKDATCINDVNGFRC-MCPEGPHHPSCYSQVNECLSSPCIHGNCTGGLSGYKCLCD 746
Query: 187 RGECGINPEAMLVTCKKTCNQCGNPGKCSDY--NRNCTKWAREGLCEKNFNFMQKNCKFS 360
G GIN E C N C N G C++ CT C+K F NC+ +
Sbjct: 747 AGWVGINCEVDKNECLS--NPCQNGGTCNNLVNGYRCT-------CKKGFKGY--NCQVN 795
Query: 361 CDACQDIGDGDNSNPNCLNSHESCDTWADRGECASNPGYMLVSCKKACNVCGKENKC--A 534
D C SNP CLN D + C Y +C+ C N C A
Sbjct: 796 IDECA-------SNP-CLNQGTCLDDVSGY-TCHCMLPYTGKNCQTVLAPCSP-NPCENA 845
Query: 535 DYRKRCPIMAKYGLCESDPRFMLQNCKESCGVCDGSSKNRPCVDANQDCAGWAASHECET 714
K P + C P + Q C C ++PC++ N C S+ CE
Sbjct: 846 AVCKEAPNFESF-TCLCAPGWQGQRCTVDVDEC----VSKPCMN-NGICHNTQGSYMCEC 899
Query: 715 NPSWMLYNCKKSCNHCRDARLESISNPNCKDLQIHCLYWANAGECQLNPGWAKQNCPFSC 894
P + +C++ N C ++NP C++ C+ N C PG+ C
Sbjct: 900 PPGFSGMDCEEDINDC-------LANP-CQN-GGSCVDKVNTFSCLCLPGFVGDKCQTDM 950
Query: 895 QHC--GGCRDLIQNCQNFANQGYCKTNEVYMNQSCKKTCGFC 1014
C C++ C ++ N C + C+ C
Sbjct: 951 NECLSEPCKN-GGTCSDYVNSYTCTCPAGFHGVHCENNIDEC 991
Score = 38.5 bits (88), Expect = 0.034
Identities = 73/354 (20%), Positives = 114/354 (32%), Gaps = 25/354 (7%)
Frame = +1
Query: 31 NQYCQEWKKSGYCEKEPNTMKEYCSK-TCGF-CESETED----PNCIDRDRNCQMWSDRG 192
NQ CQ G C N K C T G CE +D P+C++ + C
Sbjct: 1193 NQPCQN---GGTCIDLVNHFKCSCPPGTRGLLCEENIDDCAGAPHCLNGGQ-CVDRIGGY 1248
Query: 193 ECGINPEAMLVTCKKTCNQCGNPGKCSDYNRNCTKWAREGLCEKNFNFMQKNCKFSCDAC 372
C P C+ N+C + S+ + +C + C F ++C+ D C
Sbjct: 1249 SCRCLPGFAGERCEGDINECLSNPCSSEGSLDCIQLKNNYQCVCRSAFTGRHCETFLDVC 1308
Query: 373 QDIGDGDNSNPNCLNSHES--CDTWADRGECASNPGYMLVSCKKACN--VCGKENKCADY 540
CLN D C PG+ C+ +C C + +C
Sbjct: 1309 --------PQKPCLNGGTCAVASNVPDGFICRCPPGFSGARCQSSCGQVKCRRGEQCVH- 1359
Query: 541 RKRCPIMAKYGLCESDPRFMLQNCKESCGVCDGSSKNRPCVDANQDC--AGWAASHECET 714
S P N K+ C+ + PC C + C
Sbjct: 1360 ------------TASGPHCFCPNHKD----CESGCASNPCQHGG-TCYPQRQPPYYSCRC 1402
Query: 715 NPSWMLYNCKK----------SC--NHCRDARLESISNPNCKDLQIHCLYWANAGECQLN 858
+P + +C+ +C +C D + I + C H W + G+C L
Sbjct: 1403 SPPFWGSHCESYTAPTSTPPATCLSQYCADKARDGICDEACNS---HACQW-DGGDCSLT 1458
Query: 859 PGWAKQNCPFSCQHCGGCRDLIQN-CQNFANQGYCKTNEVYMNQSCKKTCGFCR 1017
NC S + C + I N C N C ++ N C++ C+
Sbjct: 1459 MEDPWANCTSSLR----CWEYINNQCDELCNTAEC----LFDNFECQRNSKTCK 1504
Score = 37.7 bits (86), Expect = 0.059
Identities = 40/175 (22%), Positives = 61/175 (34%), Gaps = 12/175 (6%)
Frame = +1
Query: 229 CKKTCNQCGNPGKCSDYNRNCTKWAREGLCEKNFNFMQKNCKFSCDACQDIGDGDNSNPN 408
C N+C PG+C + C C+ F ++C C +
Sbjct: 179 CDADINECDIPGRC-QHGGTCLNLPGSYRCQCPQRFTGQHCDSPYVPC--------APSP 229
Query: 409 CLNSHESCDTWADRGECASNPGYMLVSCKKACNVCGKENKCAD----------YRKRCPI 558
C+N T EC PG+ +C++ + C +KC + Y RCP
Sbjct: 230 CVNGGTCRQTGDFTSECHCLPGFEGSNCERNIDDC-PNHKCQNGGVCVDGVNTYNCRCPP 288
Query: 559 MAKYGLCESD-PRFMLQ-NCKESCGVCDGSSKNRPCVDANQDCAGWAASHECETN 717
C D +LQ N ++ G C + CV N GW+ +C N
Sbjct: 289 QWTGQFCTEDVDECLLQPNACQNGGTCTNRNGGYGCVCVN----GWSGD-DCSEN 338
>sp|Q14162|SREC_HUMAN Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1) Length = 830 Score = 63.9 bits (154), Expect = 8e-10 Identities = 82/360 (22%), Positives = 107/360 (29%), Gaps = 43/360 (11%) Frame = +1 Query: 55 KSGYCEKEPNTMKEYCSKTC------------------GFCESETEDPNCIDRDRNCQMW 180 K G C +P +CS C G CE T C W Sbjct: 71 KPGLCRCKPGFFGAHCSSRCPGQYWGPDCRESCPCHPHGQCEPATGACQC-----QADRW 125 Query: 181 SDRGE--CGINPEAMLVTCKKTCN--------QCGNPGKCSDYNRNCTKWAREGLCEKNF 330 R E C P C+ C P +C+ C + G C Sbjct: 126 GARCEFPCACGPHGRCDPATGVCHCEPGWWSSTCRRPCQCNTAAARCEQ--ATGACVCKP 183 Query: 331 NFMQKNCKFSCDACQDIGDGDNSNPNCLNSHESCDTWADRGECASNPGYMLVSCKKACNV 510 + + C F C NC S C+ D G CA PG+ C++ C Sbjct: 184 GWWGRRCSFRC--------------NCHGS--PCEQ--DSGRCACRPGWWGPECQQQCEC 225 Query: 511 CGKENKCADYRKRCPIMAKYGLCE--SDPRFMLQNCKESCGVCDGSSKNRPCVDANQDCA 684 A CP + CE C SCG C N PC C Sbjct: 226 VRGRCSAASGECTCPPGFRGARCELPCPAGSHGVQCAHSCGRC---KHNEPCSPDTGSC- 281 Query: 685 GWAASHECETNPSWMLYNCKKSCNHCRDARLESISNPNCKDLQIHCLYW----ANAGECQ 852 CE P W C++ C L +C+ HC + + G CQ Sbjct: 282 -----ESCE--PGWNGTQCQQPC-------LPGTFGESCEQQCPHCRHGEACEPDTGHCQ 327 Query: 853 -LNPGWAKQNCPFSC------QHCGG-CRDLIQ-NCQNFANQGYCKTNEVYMNQSCKKTC 1005 +PGW C C + CG C +Q +C G C + Y SC +C Sbjct: 328 RCDPGWLGPRCEDPCPTGTFGEDCGSTCPTCVQGSCDTVT--GDCVCSAGYWGPSCNASC 385
Score = 38.5 bits (88), Expect = 0.034
Identities = 35/162 (21%), Positives = 59/162 (36%), Gaps = 2/162 (1%)
Frame = +1
Query: 100 CSKTCGFCESETEDPNCIDRDRNCQMWSDRGECG--INPEAMLVTCKKTCNQCGNPGKCS 273
C+ +CG C+ +C+ + +C P +C++ C C + C
Sbjct: 261 CAHSCGRCKHNEPCSPDTGSCESCEPGWNGTQCQQPCLPGTFGESCEQQCPHCRHGEACE 320
Query: 274 DYNRNCTKWAREGLCEKNFNFMQKNCKFSCDACQDIGDGDNSNPNCLNSHESCDTWADRG 453
+C + C+ + + C+ C D ++ P C+ SCDT G
Sbjct: 321 PDTGHCQR------CDPGW--LGPRCEDPCPTGTFGEDCGSTCPTCVQG--SCDTVT--G 368
Query: 454 ECASNPGYMLVSCKKACNVCGKENKCADYRKRCPIMAKYGLC 579
+C + GY SC +C N C+ P GLC
Sbjct: 369 DCVCSAGYWGPSCNASCPAGFHGNNCS-----VPCECPEGLC 405
>sp|Q9IAT6|DLLC_BRARE Delta-like protein C precursor (DeltaC protein) (delC) Length = 664 Score = 63.2 bits (152), Expect = 1e-09 Identities = 84/318 (26%), Positives = 122/318 (38%), Gaps = 17/318 (5%) Frame = +1 Query: 16 CEDKPNQYCQEWKKSGYCEKEPNTMKEYCSKTCGFCESETEDPNCIDRDRNCQM-WSDR- 189 C+ N+ C K YC EP + CS+ G+CE+ E C C++ W Sbjct: 181 CDAAGNRICLPGWKGDYCT-EPICLSG-CSEENGYCEAPGE---C-----KCRIGWEGPL 230 Query: 190 -GECGINPEAMLVTCKKTCNQCGNPGKCSDYNRNCTKWAREGLCEKNFNFMQKNCKFSCD 366 EC +P + TCNQ P +C+ C + C ++ NF + D Sbjct: 231 CDECTRHPGCL----HGTCNQ---PFQCT-----CKEGWGGLFCNEDLNFCTNHKPCRND 278 Query: 367 A-CQDIGDGDNSNPNCLN----SHESCDTWADRGECASNPGYMLVSCKKACNVCGKENKC 531 A C + G G + C+ S ++C+ + EC SNP CK + +EN Sbjct: 279 ATCTNTGQGSYT---CICKPGFSGKNCEI--ETNECDSNP------CKNGGSCNDQEN-- 325 Query: 532 ADYRKRCPIMAKYGLCESDPRFMLQNCKESCGVC-DGSSKNRPCVDANQDCAGWAASHEC 708 DY CP YG +NC+ S C DG PC + + S+ C Sbjct: 326 -DYTCTCP-QGFYG----------KNCEVSAMTCADG-----PCFNGGTCMEKGSGSYSC 368 Query: 709 ETNPSWMLYNCKKSCNHCRDARLESISNPNCKDLQIHCLYWANAGECQLNPGWAKQNCPF 888 P +M NC+K + C S+P Q CL N C+ PG+ C Sbjct: 369 RCPPGYMGSNCEKKIDRCS-------SDPCANGGQ--CLDLGNKATCRCRPGFTGSRCET 419 Query: 889 S--------CQHCGGCRD 918 + CQ+ G C D Sbjct: 420 NIDDCSSNPCQNAGTCVD 437
Score = 38.9 bits (89), Expect = 0.026
Identities = 43/171 (25%), Positives = 52/171 (30%), Gaps = 6/171 (3%)
Frame = +1
Query: 1 IVASVCEDKP---NQYCQEWKKSGY-CEKEPNTMKEYCSKTCGFCESE--TEDPNCIDRD 162
+ A C D P C E Y C P M C K C S+ C+D
Sbjct: 342 VSAMTCADGPCFNGGTCMEKGSGSYSCRCPPGYMGSNCEKKIDRCSSDPCANGGQCLDLG 401
Query: 163 RNCQMWSDRGECGINPEAMLVTCKKTCNQCGNPGKCSDYNRNCTKWAREGLCEKNFNFMQ 342
G G E + C N C N G C D T C F
Sbjct: 402 NKATCRCRPGFTGSRCETNIDDCSS--NPCQNAGTCVDGINGYT-------CTCTLGFSG 452
Query: 343 KNCKFSCDACQDIGDGDNSNPNCLNSHESCDTWADRGECASNPGYMLVSCK 495
K+C+ DAC S C N +C T C G+M C+
Sbjct: 453 KDCRVRSDAC--------SFMPCQNG-GTCYTHFSGPVCQCPAGFMGTQCE 494
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 139,004,359
Number of Sequences: 369166
Number of extensions: 3295196
Number of successful extensions: 10843
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8658
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10129
length of database: 68,354,980
effective HSP length: 112
effective length of database: 47,664,660
effective search space used: 12059158980
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail