DrC_00878
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
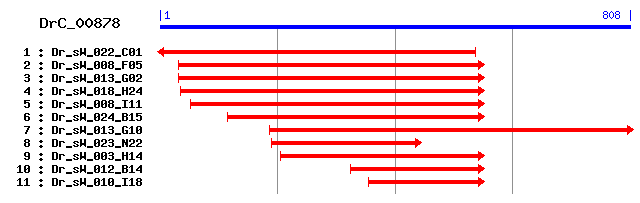
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00878 (808 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O60830|TI17B_HUMAN Mitochondrial import inner membrane t... 130 3e-30 sp|Q99595|TI17A_HUMAN Mitochondrial import inner membrane t... 129 7e-30 sp|Q9Z0V8|TI17A_MOUSE Mitochondrial import inner membrane t... 128 2e-29 sp|Q9VNA0|TI17A_DROME Probable mitochondrial import inner m... 128 2e-29 sp|Q9Z0V7|TI17B_MOUSE Mitochondrial import inner membrane t... 127 4e-29 sp|O35092|TI17A_RAT Mitochondrial import inner membrane tra... 127 4e-29 sp|Q9VGA2|TI17C_DROME Probable mitochondrial import inner m... 122 1e-27 sp|P39515|TIM17_YEAST Mitochondrial import inner membrane t... 118 2e-26 sp|P59670|TIM17_NEUCR Mitochondrial import inner membrane t... 118 2e-26 sp|O44477|TIM17_CAEEL Probable mitochondrial import inner m... 117 5e-26
>sp|O60830|TI17B_HUMAN Mitochondrial import inner membrane translocase subunit Tim17 B (JM3) Length = 172 Score = 130 bits (328), Expect = 3e-30 Identities = 64/139 (46%), Positives = 86/139 (61%), Gaps = 1/139 (0%) Frame = +2 Query: 20 MDAY-RDPCPWRIFSDXXXXXXXXXXXXXLFNFVAGFKDAPSGLSKRLISAATRCKIRSP 196 M+ Y R+PCPWRI D +F + GF++AP G+ RL +A +IR+P Sbjct: 1 MEEYAREPCPWRIVDDCGGAFTMGVIGGGVFQAIKGFRNAPVGIRHRLRGSANAVRIRAP 60 Query: 197 VLGGSFAMWGGTFSCVDCALIYGRKKEDPINSILAGGITGGVLAIRGGWSQVAGQAFVGA 376 +GGSFA+WGG FS +DC L+ R KEDP NSI +G +TG VLA R G + G A +G Sbjct: 61 QIGGSFAVWGGLFSTIDCGLVRLRGKEDPWNSITSGALTGAVLAARSGPLAMVGSAMMGG 120 Query: 377 VLLGIIEGCGILFQRFSTQ 433 +LL +IEG GIL R++ Q Sbjct: 121 ILLALIEGVGILLTRYTAQ 139
>sp|Q99595|TI17A_HUMAN Mitochondrial import inner membrane translocase subunit Tim17 A (Inner membrane preprotein translocase Tim17a) Length = 171 Score = 129 bits (325), Expect = 7e-30 Identities = 62/138 (44%), Positives = 87/138 (63%), Gaps = 1/138 (0%) Frame = +2 Query: 20 MDAY-RDPCPWRIFSDXXXXXXXXXXXXXLFNFVAGFKDAPSGLSKRLISAATRCKIRSP 196 M+ Y R+PCPWRI D +F + GF+++P G++ RL + T K R+P Sbjct: 1 MEEYAREPCPWRIVDDCGGAFTMGTIGGGIFQAIKGFRNSPVGVNHRLRGSLTAIKTRAP 60 Query: 197 VLGGSFAMWGGTFSCVDCALIYGRKKEDPINSILAGGITGGVLAIRGGWSQVAGQAFVGA 376 LGGSFA+WGG FS +DC+++ R KEDP NSI +G +TG +LA R G + G A +G Sbjct: 61 QLGGSFAVWGGLFSMIDCSMVQVRGKEDPWNSITSGALTGAILAARNGPVAMVGSAAMGG 120 Query: 377 VLLGIIEGCGILFQRFST 430 +LL +IEG GIL RF++ Sbjct: 121 ILLALIEGAGILLTRFAS 138
>sp|Q9Z0V8|TI17A_MOUSE Mitochondrial import inner membrane translocase subunit Tim17 A (Inner membrane preprotein translocase Tim17a) Length = 171 Score = 128 bits (321), Expect = 2e-29 Identities = 62/138 (44%), Positives = 86/138 (62%), Gaps = 1/138 (0%) Frame = +2 Query: 20 MDAY-RDPCPWRIFSDXXXXXXXXXXXXXLFNFVAGFKDAPSGLSKRLISAATRCKIRSP 196 M+ Y R+PCPWRI D +F GF+++P G++ RL + T K R+P Sbjct: 1 MEEYAREPCPWRIVDDCGGAFTMGTIGGGIFQAFKGFRNSPVGINHRLRGSLTAIKTRAP 60 Query: 197 VLGGSFAMWGGTFSCVDCALIYGRKKEDPINSILAGGITGGVLAIRGGWSQVAGQAFVGA 376 LGGSFA+WGG FS +DC+++ R KEDP NSI +G +TG +LA R G + G A +G Sbjct: 61 QLGGSFAVWGGLFSTIDCSMVQIRGKEDPWNSITSGALTGAILAARNGPVAMVGSAAMGG 120 Query: 377 VLLGIIEGCGILFQRFST 430 +LL +IEG GIL RF++ Sbjct: 121 ILLALIEGAGILLTRFAS 138
>sp|Q9VNA0|TI17A_DROME Probable mitochondrial import inner membrane translocase subunit Tim17 1 Length = 179 Score = 128 bits (321), Expect = 2e-29 Identities = 63/156 (40%), Positives = 89/156 (57%) Frame = +2 Query: 32 RDPCPWRIFSDXXXXXXXXXXXXXLFNFVAGFKDAPSGLSKRLISAATRCKIRSPVLGGS 211 R+PCP+RI D F + GF++APSGL RL + RS ++GG+ Sbjct: 6 REPCPFRIVEDCGGAFAMGALGGGAFQAIKGFRNAPSGLGYRLSGGLAAVRARSGLVGGN 65 Query: 212 FAMWGGTFSCVDCALIYGRKKEDPINSILAGGITGGVLAIRGGWSQVAGQAFVGAVLLGI 391 FA+WG TFS +DC+L+Y RKKEDP N+I++G TGG+LA R G + + A VG LL + Sbjct: 66 FAVWGATFSAIDCSLVYFRKKEDPWNAIISGATTGGILAARTGLTSMLSSALVGGALLAL 125 Query: 392 IEGCGILFQRFSTQAIVEESMPDNNQTISHQSLGLQ 499 IEG GI+ +S + + S + Q + L Q Sbjct: 126 IEGVGIVVSHYSADSYRQVSPVERQQRYKQELLRQQ 161
>sp|Q9Z0V7|TI17B_MOUSE Mitochondrial import inner membrane translocase subunit Tim17 B Length = 172 Score = 127 bits (319), Expect = 4e-29 Identities = 62/139 (44%), Positives = 84/139 (60%), Gaps = 1/139 (0%) Frame = +2 Query: 20 MDAY-RDPCPWRIFSDXXXXXXXXXXXXXLFNFVAGFKDAPSGLSKRLISAATRCKIRSP 196 M+ Y R+PCPWRI D +F + GF++AP G+ R + +IR+P Sbjct: 1 MEEYAREPCPWRIVDDCGGAFTMGVIGGGVFQAIKGFRNAPVGIRHRFRGSVNAVRIRAP 60 Query: 197 VLGGSFAMWGGTFSCVDCALIYGRKKEDPINSILAGGITGGVLAIRGGWSQVAGQAFVGA 376 +GGSFA+WGG FS +DC L+ R KEDP NSI +G +TG VLA R G + G A +G Sbjct: 61 QIGGSFAVWGGLFSTIDCGLVRLRGKEDPWNSISSGALTGAVLAARSGPLAMVGSAMMGG 120 Query: 377 VLLGIIEGCGILFQRFSTQ 433 +LL +IEG GIL R++ Q Sbjct: 121 ILLALIEGVGILLTRYTAQ 139
>sp|O35092|TI17A_RAT Mitochondrial import inner membrane translocase subunit Tim17 A (Inner membrane preprotein translocase Tim17a) Length = 171 Score = 127 bits (319), Expect = 4e-29 Identities = 62/138 (44%), Positives = 85/138 (61%), Gaps = 1/138 (0%) Frame = +2 Query: 20 MDAY-RDPCPWRIFSDXXXXXXXXXXXXXLFNFVAGFKDAPSGLSKRLISAATRCKIRSP 196 M+ Y R+PCPWRI D +F GF+++P G++ RL + T K R+P Sbjct: 1 MEEYAREPCPWRIVDDCGGAFTMGTIGGGIFQAFKGFRNSPVGVNHRLRGSLTAIKTRAP 60 Query: 197 VLGGSFAMWGGTFSCVDCALIYGRKKEDPINSILAGGITGGVLAIRGGWSQVAGQAFVGA 376 LGGSFA+WGG FS +DC ++ R KEDP NSI +G +TG +LA R G + G A +G Sbjct: 61 QLGGSFAVWGGLFSTIDCGMVQIRGKEDPWNSITSGALTGAILAARNGPVAMVGSAAMGG 120 Query: 377 VLLGIIEGCGILFQRFST 430 +LL +IEG GIL RF++ Sbjct: 121 ILLALIEGAGILLTRFAS 138
>sp|Q9VGA2|TI17C_DROME Probable mitochondrial import inner membrane translocase subunit Tim17 3 Length = 222 Score = 122 bits (306), Expect = 1e-27 Identities = 60/129 (46%), Positives = 80/129 (62%) Frame = +2 Query: 20 MDAYRDPCPWRIFSDXXXXXXXXXXXXXLFNFVAGFKDAPSGLSKRLISAATRCKIRSPV 199 M+ R PCP RI D LF F+ GF++AP+GL +RL K+R+P Sbjct: 1 MEYNRQPCPIRIVEDCGCAFMMGTIGGSLFEFLKGFRNAPTGLQRRLYGGIDLVKMRTPS 60 Query: 200 LGGSFAMWGGTFSCVDCALIYGRKKEDPINSILAGGITGGVLAIRGGWSQVAGQAFVGAV 379 + GSFA+WG TFS VDCAL++ R++ED NSIL+G TGG+LA R G +A A VG + Sbjct: 61 IAGSFAVWGATFSTVDCALVHYRQREDAWNSILSGAATGGILAARNGIRAMANSALVGCL 120 Query: 380 LLGIIEGCG 406 +L +IEG G Sbjct: 121 VLAMIEGAG 129
>sp|P39515|TIM17_YEAST Mitochondrial import inner membrane translocase subunit TIM17 (Mitochondrial protein import protein 2) (Mitochondrial inner membrane protein MIM17) Length = 158 Score = 118 bits (296), Expect = 2e-26 Identities = 59/135 (43%), Positives = 83/135 (61%) Frame = +2 Query: 23 DAYRDPCPWRIFSDXXXXXXXXXXXXXLFNFVAGFKDAPSGLSKRLISAATRCKIRSPVL 202 D RDPCP I +D +++ + GF+++P G +R A + K R+PVL Sbjct: 4 DHSRDPCPIVILNDFGGAFAMGAIGGVVWHGIKGFRNSPLG--ERGSGAMSAIKARAPVL 61 Query: 203 GGSFAMWGGTFSCVDCALIYGRKKEDPINSILAGGITGGVLAIRGGWSQVAGQAFVGAVL 382 GG+F +WGG FS DCA+ RK+EDP N+I+AG TGG LA+RGGW + A L Sbjct: 62 GGNFGVWGGLFSTFDCAVKAVRKREDPWNAIIAGFFTGGALAVRGGWRHTRNSSITCACL 121 Query: 383 LGIIEGCGILFQRFS 427 LG+IEG G++FQR++ Sbjct: 122 LGVIEGVGLMFQRYA 136
>sp|P59670|TIM17_NEUCR Mitochondrial import inner membrane translocase subunit tim-17 Length = 155 Score = 118 bits (295), Expect = 2e-26 Identities = 68/155 (43%), Positives = 90/155 (58%), Gaps = 3/155 (1%) Frame = +2 Query: 20 MDAYRDPCPWRIFSDXXXXXXXXXXXXXLFNFVAGFKDAPSGLSKRLISAATRCKIRSPV 199 MD RDPCPW I +D +++ + GF+++P G +R I A T K+R+P Sbjct: 1 MDHTRDPCPWVILNDFGGAFAMGAIGGTIWHGIKGFRNSPYG--ERRIGAITAIKMRAPA 58 Query: 200 LGGSFAMWGGTFSCVDCALIYGRK-KEDPINSILAGGITGGVLAIRGGWSQVAGQAFVGA 376 LGG+F +WGG FS DCA+ R KEDP NSILAG TGG LA+RGG+ A A Sbjct: 59 LGGNFGVWGGLFSTFDCAIKGLRNHKEDPWNSILAGFFTGGALAVRGGYKAARNGAIGCA 118 Query: 377 VLLGIIEGCGILFQRFSTQAIVEE--SMPDNNQTI 475 VLL +IEG GI FQ+ A E + P +N+ + Sbjct: 119 VLLAVIEGVGIGFQKMLAGATKLEAPAPPPSNEKV 153
>sp|O44477|TIM17_CAEEL Probable mitochondrial import inner membrane translocase subunit Tim17 Length = 181 Score = 117 bits (292), Expect = 5e-26 Identities = 64/159 (40%), Positives = 94/159 (59%), Gaps = 1/159 (0%) Frame = +2 Query: 20 MDAY-RDPCPWRIFSDXXXXXXXXXXXXXLFNFVAGFKDAPSGLSKRLISAATRCKIRSP 196 M+ Y R+PCP+RI D +F G+K+A G K+L+ ++RS Sbjct: 1 MEEYTREPCPYRIGDDIGSAFAMGLVGGSIFQAFGGYKNAAKG--KKLVGMMREVRMRST 58 Query: 197 VLGGSFAMWGGTFSCVDCALIYGRKKEDPINSILAGGITGGVLAIRGGWSQVAGQAFVGA 376 + G FA WGG FS +DC L+ RKKEDPINSI++GG+TG +LAIR G +AG A +G+ Sbjct: 59 LTGVQFAAWGGMFSTIDCCLVAIRKKEDPINSIVSGGLTGALLAIRSGPKVMAGSAILGS 118 Query: 377 VLLGIIEGCGILFQRFSTQAIVEESMPDNNQTISHQSLG 493 V+L +IEG G++ R+ A+++ + P +SLG Sbjct: 119 VILAMIEGVGLVTTRW-MGAMMDPTQPPPEALDDPRSLG 156
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 87,799,326
Number of Sequences: 369166
Number of extensions: 1791429
Number of successful extensions: 4850
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 4538
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4811
length of database: 68,354,980
effective HSP length: 109
effective length of database: 48,218,865
effective search space used: 7666799535
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail