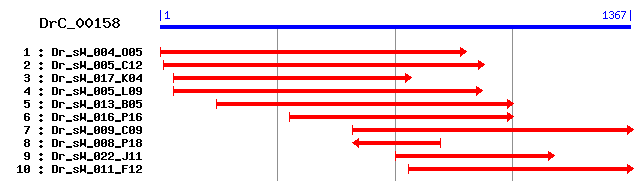
DrC_00158
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00158 (1367 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O08730|GLYG_RAT Glycogenin-1 326 1e-88 sp|Q9R062|GLYG_MOUSE Glycogenin-1 323 5e-88 sp|P46976|GLYG_HUMAN Glycogenin-1 321 4e-87 sp|P13280|GLYG_RABIT Glycogenin-1 320 8e-87 sp|O15488|GLYG2_HUMAN Glycogenin-2 (GN-2) (GN2) 282 2e-75 sp|P47011|GLG2_YEAST Glycogen synthesis initiator protein GLG2 123 1e-27 sp|P36143|GLG1_YEAST Glycogen synthesis initiator protein GLG1 84 1e-15 sp|Q9Y761|GNT1A_KLULA Glucose N-acetyltransferase 1-A (N-ac... 42 0.004 sp|Q6CT96|GNT1B_KLULA Glucose N-acetyltransferase 1-B (N-ac... 37 0.13 sp|Q09680|YA0C_SCHPO Hypothetical protein C5H10.12c in chro... 36 0.30
>sp|O08730|GLYG_RAT Glycogenin-1 Length = 333 Score = 326 bits (835), Expect = 1e-88 Identities = 167/317 (52%), Positives = 208/317 (65%), Gaps = 14/317 (4%) Frame = +1 Query: 40 ALGALTLAMSLKHSGTQKELVIMITDNISEAMRNVLSQVFNHIEMVNVLDSNDVINLGLL 219 A GAL L SLK T + V++ + +S++MR VL VF+ + MV+VLDS D +L L+ Sbjct: 16 AKGALVLGSSLKQHRTTRRTVVLASPQVSDSMRKVLETVFDEVIMVDVLDSGDSAHLTLM 75 Query: 220 ERPELGITFTKLHCWRLVQYSKCVFMDADTLVIQNIDDLFEREELSAAPDPGWPDCFNSG 399 +RPELGIT TKLHCW L QYSKCVFMDADTLV+ NIDDLFEREELSAAPDPGWPDCFNSG Sbjct: 76 KRPELGITLTKLHCWSLTQYSKCVFMDADTLVLSNIDDLFEREELSAAPDPGWPDCFNSG 135 Query: 400 VFVYKPSIDTYVELLQFAIDKGSFDGGDQGLLNMFFSNWSTKDIKHHLPFVYNCVSQAFY 579 VFVY+PSI+TY +LL A ++GSFDGGDQGLLN +FS W+T DI HLPFVYN S + Y Sbjct: 136 VFVYQPSIETYNQLLHLASEQGSFDGGDQGLLNTYFSGWATTDITKHLPFVYNLSSLSIY 195 Query: 580 SYLPAYTHFRADIKVLHFIGPHKPWHHTFDSDTGTV--IFQEGCGQNYESLQLWWTTFIA 753 SYLPA+ F + KV+HF+G KPW++T++ T +V Q+ + E L LWW TF Sbjct: 196 SYLPAFKAFGKNAKVVHFLGRTKPWNYTYNPQTKSVKCESQDPIVSHPEFLNLWWDTFTT 255 Query: 754 YTKPLLHDE-----------MGGVCGRMASLDVSSFVP-HEGKFSPKSHQQAWEHGIIDY 897 PLL M V G ++ L P + S + ++ WE G DY Sbjct: 256 NVLPLLQHHGLVKDAGSYLMMEHVTGALSDLSFGEAPPASQPSLSSEERKERWEQGQADY 315 Query: 898 RGLDRFENIKMHLDSKL 948 G D F+NIK LD+ L Sbjct: 316 MGADSFDNIKRKLDTYL 332
>sp|Q9R062|GLYG_MOUSE Glycogenin-1 Length = 333 Score = 323 bits (829), Expect = 5e-88 Identities = 166/317 (52%), Positives = 208/317 (65%), Gaps = 14/317 (4%) Frame = +1 Query: 40 ALGALTLAMSLKHSGTQKELVIMITDNISEAMRNVLSQVFNHIEMVNVLDSNDVINLGLL 219 A GAL L SLK T + +V++ + +S++MR VL VF+ + MV+VLDS D +L L+ Sbjct: 16 AKGALVLGSSLKQHRTTRRMVVLTSPQVSDSMRKVLETVFDDVIMVDVLDSGDSAHLTLM 75 Query: 220 ERPELGITFTKLHCWRLVQYSKCVFMDADTLVIQNIDDLFEREELSAAPDPGWPDCFNSG 399 +RPELGIT TKLHCW L QYSKCVFMDADTLV+ NIDDLFEREELSAAPDPGWPDCFNSG Sbjct: 76 KRPELGITLTKLHCWSLTQYSKCVFMDADTLVLSNIDDLFEREELSAAPDPGWPDCFNSG 135 Query: 400 VFVYKPSIDTYVELLQFAIDKGSFDGGDQGLLNMFFSNWSTKDIKHHLPFVYNCVSQAFY 579 VFVY+PSI+TY +LL A ++GSFDGGDQGLLN +FS W+T DI HLPFVYN S + Y Sbjct: 136 VFVYQPSIETYNQLLHLASEQGSFDGGDQGLLNTYFSGWATTDITKHLPFVYNLSSISIY 195 Query: 580 SYLPAYTHFRADIKVLHFIGPHKPWHHTFDSDTGTV--IFQEGCGQNYESLQLWWTTFIA 753 SYLPA+ F + KV+HF+G KPW++T++ T +V Q+ + E L LWW TF Sbjct: 196 SYLPAFKAFGKNAKVVHFLGRTKPWNYTYNPQTKSVNCDSQDPTVSHPEFLNLWWDTFTT 255 Query: 754 YTKPLLHDE-----------MGGVCGRMASLDVSSF-VPHEGKFSPKSHQQAWEHGIIDY 897 PLL M V G ++ L + S + ++ WE G DY Sbjct: 256 NVLPLLQHHGLVKDASSYLMMEHVSGALSDLSFGEAPAAPQPSMSSEERKERWEQGQADY 315 Query: 898 RGLDRFENIKMHLDSKL 948 G D F+NIK LD+ L Sbjct: 316 MGADSFDNIKRKLDTYL 332
>sp|P46976|GLYG_HUMAN Glycogenin-1 Length = 350 Score = 321 bits (822), Expect = 4e-87 Identities = 168/335 (50%), Positives = 210/335 (62%), Gaps = 32/335 (9%) Frame = +1 Query: 40 ALGALTLAMSLKHSGTQKELVIMITDNISEAMRNVLSQVFNHIEMVNVLDSNDVINLGLL 219 A GAL L SLK T + LV++ T +S++MR VL VF+ + MV+VLDS D +L L+ Sbjct: 16 AKGALVLGSSLKQHRTTRRLVVLATPQVSDSMRKVLETVFDEVIMVDVLDSGDSAHLTLM 75 Query: 220 ERPELGITFTKLHCWRLVQYSKCVFMDADTLVIQNIDDLFEREELSAAPDPGWPDCFNSG 399 +RPELG+T TKLHCW L QYSKCVFMDADTLV+ NIDDLF+REELSAAPDPGWPDCFNSG Sbjct: 76 KRPELGVTLTKLHCWSLTQYSKCVFMDADTLVLANIDDLFDREELSAAPDPGWPDCFNSG 135 Query: 400 VFVYKPSIDTYVELLQFAIDKGSFDGGDQGLLNMFFSNWSTKDIKHHLPFVYNCVSQAFY 579 VFVY+PS++TY +LL A ++GSFDGGDQG+LN FFS+W+T DI+ HLPF+YN S + Y Sbjct: 136 VFVYQPSVETYNQLLHLASEQGSFDGGDQGILNTFFSSWATTDIRKHLPFIYNLSSISIY 195 Query: 580 SYLPAYTHFRADIKVLHFIGPHKPWHHTFDSDTGTVIFQEGCGQNY---ESLQLWWTTFI 750 SYLPA+ F A KV+HF+G KPW++T+D T +V E N E L LWW F Sbjct: 196 SYLPAFKVFGASAKVVHFLGRVKPWNYTYDPKTKSV-KSEAHDPNMTHPEFLILWWNIFT 254 Query: 751 AYTKPLLHD-------------------EMGGVCGRMASLDVSSFVPH----------EG 843 PLL + CG DVS + H + Sbjct: 255 TNVLPLLQQFGLVKDTCSYVNVLSDLVYTLAFSCGFCRKEDVSGAISHLSLGEIPAMAQP 314 Query: 844 KFSPKSHQQAWEHGIIDYRGLDRFENIKMHLDSKL 948 S + ++ WE G DY G D F+NIK LD+ L Sbjct: 315 FVSSEERKERWEQGQADYMGADSFDNIKRKLDTYL 349
>sp|P13280|GLYG_RABIT Glycogenin-1 Length = 333 Score = 320 bits (819), Expect = 8e-87 Identities = 161/317 (50%), Positives = 208/317 (65%), Gaps = 14/317 (4%) Frame = +1 Query: 40 ALGALTLAMSLKHSGTQKELVIMITDNISEAMRNVLSQVFNHIEMVNVLDSNDVINLGLL 219 A GAL L SLK T + L ++ T +S+ MR L VF+ + V++LDS D +L L+ Sbjct: 16 AKGALVLGSSLKQHRTSRRLAVLTTPQVSDTMRKALEIVFDEVITVDILDSGDSAHLTLM 75 Query: 220 ERPELGITFTKLHCWRLVQYSKCVFMDADTLVIQNIDDLFEREELSAAPDPGWPDCFNSG 399 +RPELG+T TKLHCW L QYSKCVFMDADTLV+ NIDDLFEREELSAAPDPGWPDCFNSG Sbjct: 76 KRPELGVTLTKLHCWSLTQYSKCVFMDADTLVLANIDDLFEREELSAAPDPGWPDCFNSG 135 Query: 400 VFVYKPSIDTYVELLQFAIDKGSFDGGDQGLLNMFFSNWSTKDIKHHLPFVYNCVSQAFY 579 VFVY+PS++TY +LL A ++GSFDGGDQGLLN FF++W+T DI+ HLPF+YN S + Y Sbjct: 136 VFVYQPSVETYNQLLHVASEQGSFDGGDQGLLNTFFNSWATTDIRKHLPFIYNLSSISIY 195 Query: 580 SYLPAYTHFRADIKVLHFIGPHKPWHHTFDSDTGTVIFQ--EGCGQNYESLQLWWTTFIA 753 SYLPA+ F A+ KV+HF+G KPW++T+D+ T +V + + + + L +WW F Sbjct: 196 SYLPAFKAFGANAKVVHFLGQTKPWNYTYDTKTKSVRSEGHDPTMTHPQFLNVWWDIFTT 255 Query: 754 YTKPLLHD--EMGGVCGRMASLDVSSFVPH----------EGKFSPKSHQQAWEHGIIDY 897 PLL + C DVS V H + S + ++ WE G DY Sbjct: 256 SVVPLLQQFGLVQDTCSYQHVEDVSGAVSHLSLGETPATTQPFVSSEERKERWEQGQADY 315 Query: 898 RGLDRFENIKMHLDSKL 948 G D F+NIK LD+ L Sbjct: 316 MGADSFDNIKKKLDTYL 332
>sp|O15488|GLYG2_HUMAN Glycogenin-2 (GN-2) (GN2) Length = 501 Score = 282 bits (721), Expect = 2e-75 Identities = 135/275 (49%), Positives = 179/275 (65%), Gaps = 20/275 (7%) Frame = +1 Query: 46 GALTLAMSLKHSGTQKELVIMITDNISEAMRNVLSQVFNHIEMVNVLDSNDVINLGLLER 225 GAL L SL+ ++LV++IT +S +R +LS+VF+ + VN++DS D I+L L+R Sbjct: 51 GALVLGQSLRRHRLTRKLVVLITPQVSSLLRVILSKVFDEVIEVNLIDSADYIHLAFLKR 110 Query: 226 PELGITFTKLHCWRLVQYSKCVFMDADTLVIQNIDDLFEREELSAAPDPGWPDCFNSGVF 405 PELG+T TKLHCW L YSKCVF+DADTLV+ N+D+LF+R E SAAPDPGWPDCFNSGVF Sbjct: 111 PELGLTLTKLHCWTLTHYSKCVFLDADTLVLSNVDELFDRGEFSAAPDPGWPDCFNSGVF 170 Query: 406 VYKPSIDTYVELLQFAIDKGSFDGGDQGLLNMFFSNWSTKDIKHHLPFVYNCVSQAFYSY 585 V++PS+ T+ LLQ A++ GSFDG DQGLLN FF NWST DI HLPF+YN S Y+Y Sbjct: 171 VFQPSLHTHKLLLQHAMEHGSFDGADQGLLNSFFRNWSTTDIHKHLPFIYNLSSNTMYTY 230 Query: 586 LPAYTHFRADIKVLHFIGPHKPWHHTFDSDTGTVIFQEGCGQNYES---LQLWWTTFIAY 756 PA+ F + KV+HF+G KPW++ ++ +G+V+ Q + L LWWT + Sbjct: 231 SPAFKQFGSSAKVVHFLGSMKPWNYKYNPQSGSVLEQGSVSSSQHQAAFLHLWWTVYQNN 290 Query: 757 TKP-----------------LLHDEMGGVCGRMAS 810 P L H ++GG C AS Sbjct: 291 VLPLYKSVQAGEARASPGHTLCHSDVGGPCADSAS 325
Score = 33.5 bits (75), Expect = 1.5
Identities = 14/32 (43%), Positives = 19/32 (59%)
Frame = +1
Query: 844 KFSPKSHQQAWEHGIIDYRGLDRFENIKMHLD 939
+ SP+ ++ WE G IDY G D F I+ LD
Sbjct: 466 ELSPEEERRKWEEGRIDYMGKDAFARIQEKLD 497
>sp|P47011|GLG2_YEAST Glycogen synthesis initiator protein GLG2 Length = 380 Score = 123 bits (309), Expect = 1e-27 Identities = 88/259 (33%), Positives = 124/259 (47%), Gaps = 40/259 (15%) Frame = +1 Query: 19 LLQMMNIALGALTLAMSL----KHSGTQKELVIMIT-------DNISEAMRNVLSQVFNH 165 LL + GALTLA L KH+ + E+ + + D ++ +F Sbjct: 10 LLYSRDYLPGALTLAYQLQKLLKHAVVEDEITLCLLIEKKLFGDEFKPQEIALIRSLFKE 69 Query: 166 IEMVNVLDSNDV------INLGLLERPELGITFTKLHCWRLVQYSKCVFMDADTLVIQNI 327 I ++ L + NL LL+RPEL T K W LVQ+ + +F+DADTL + Sbjct: 70 IIIIEPLKDQEKSIEKNKANLELLKRPELSHTLLKARLWELVQFDQVLFLDADTLPLNK- 128 Query: 328 DDLFE---------REELSAAPDPGWPDCFNSGVFVYKPSIDTYVELLQFAIDKGSFDGG 480 + FE R +++A PD GWPD FN+GV + P +D L F I S DG Sbjct: 129 -EFFEILRLYPEQTRFQIAAVPDIGWPDMFNTGVLLLIPDLDMATSLQDFLIKTVSIDGA 187 Query: 481 DQGLLNMFFS---NWSTKDIKH---------HLPFVYNCVSQAF-YSYLPAYTHFRADIK 621 DQG+ N FF+ N+S K++ H LPF YN + Y PA F+ I+ Sbjct: 188 DQGIFNQFFNPICNYS-KEVLHKVSPLMEWIRLPFTYNVTMPNYGYQSSPAMNFFQQHIR 246 Query: 622 VLHFIGPHKPW-HHTFDSD 675 ++HFIG KPW +T D D Sbjct: 247 LIHFIGTFKPWSRNTTDYD 265
>sp|P36143|GLG1_YEAST Glycogen synthesis initiator protein GLG1 Length = 480 Score = 83.6 bits (205), Expect = 1e-15 Identities = 45/112 (40%), Positives = 59/112 (52%), Gaps = 10/112 (8%) Frame = +1 Query: 349 ELSAAPDPGWPDCFNSGVFVYKPSIDTYVELLQFAIDKGSFDGGDQGLLNMFFS-NWSTK 525 ++ A D GWPD FNSGV + P DT L + + S DG DQG+LN FF+ N T Sbjct: 8 QVGAIADIGWPDMFNSGVMMLIPDADTASVLQNYIFENTSIDGSDQGILNQFFNQNCCTD 67 Query: 526 DIKH--------HLPFVYN-CVSQAFYSYLPAYTHFRADIKVLHFIGPHKPW 654 ++ L F YN + Y PA +F+ IK++HFIG HKPW Sbjct: 68 ELVKDSFSREWVQLSFTYNVTIPNLGYQSSPAMNYFKPSIKLIHFIGKHKPW 119
>sp|Q9Y761|GNT1A_KLULA Glucose N-acetyltransferase 1-A (N-acetylglucosaminyltransferase A) Length = 460 Score = 42.0 bits (97), Expect = 0.004 Identities = 25/95 (26%), Positives = 51/95 (53%), Gaps = 5/95 (5%) Frame = +1 Query: 70 LKHSGTQKELVIMITDNISEAMRNVLSQVFNHIEMVNVLDSNDVI----NLGLLERPELG 237 L SGTQ +LV+++ ++E + V + + N ++ N+ L + Sbjct: 118 LHESGTQAKLVMLVAKELTELPED--DSVTRMLAQFKEISDNCIVKPVENIVLSQGSAQW 175 Query: 238 IT-FTKLHCWRLVQYSKCVFMDADTLVIQNIDDLF 339 +T TKL + +V+Y + V+ D+D+++ +N+D+LF Sbjct: 176 MTSMTKLRVFGMVEYKRIVYFDSDSIITRNMDELF 210
>sp|Q6CT96|GNT1B_KLULA Glucose N-acetyltransferase 1-B (N-acetylglucosaminyltransferase B) Length = 453 Score = 37.0 bits (84), Expect = 0.13 Identities = 25/102 (24%), Positives = 48/102 (47%), Gaps = 12/102 (11%) Frame = +1 Query: 70 LKHSGTQKELVIMITDNIS---------EAMRNVLSQVFNHI---EMVNVLDSNDVINLG 213 L SG++ +L+ ++TD + EA+ N + V + + E+ +V+ ND Sbjct: 107 LNDSGSKAKLLALVTDTLVNKSKENKEVEALLNKIKSVSDRVAVTEVGSVIQPND----- 161 Query: 214 LLERPELGITFTKLHCWRLVQYSKCVFMDADTLVIQNIDDLF 339 + TKL + L Y + ++MD D ++ +D+LF Sbjct: 162 ---HTPWSKSLTKLAIFNLTDYERIIYMDNDAIIHDKMDELF 200
>sp|Q09680|YA0C_SCHPO Hypothetical protein C5H10.12c in chromosome I Length = 371 Score = 35.8 bits (81), Expect = 0.30 Identities = 13/33 (39%), Positives = 24/33 (72%) Frame = +1 Query: 244 FTKLHCWRLVQYSKCVFMDADTLVIQNIDDLFE 342 F+KL + +Q+ K +D+D L+++NIDD+F+ Sbjct: 161 FSKLRIFEQIQFDKICVIDSDILIMKNIDDIFD 193
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 146,823,231
Number of Sequences: 369166
Number of extensions: 3003293
Number of successful extensions: 8349
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7083
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8077
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 16127659790
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail