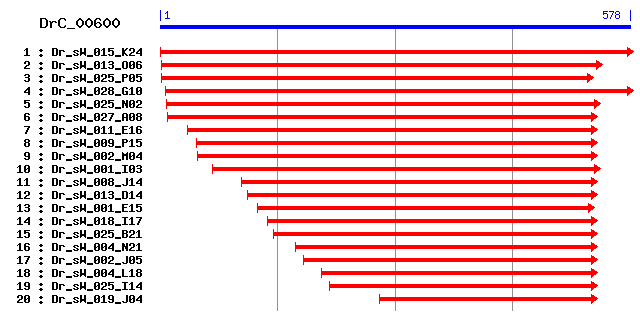
DrC_00600
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00600 (578 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P32773|TOA1_YEAST Transcription initiation factor IIA la... 38 0.021 sp|Q9N4M4|ANC1_CAEEL Nuclear anchorage protein 1 (Anchorage... 38 0.021 sp|P25980|UBF1B_XENLA Nucleolar transcription factor 1-B (U... 36 0.079 sp|P36044|MNN4_YEAST Protein MNN4 34 0.30 sp|P24788|CD2L1_MOUSE PITSLRE serine/threonine-protein kina... 33 0.39 sp|Q03101|CYAG_DICDI Adenylate cyclase, germination specifi... 33 0.39 sp|Q6FSW2|RAD52_CANGA DNA repair and recombination protein ... 33 0.51 sp|Q6DRL5|MBB1A_BRARE Myb-binding protein 1A-like protein 33 0.67 sp|Q960E8|TF2H1_DROME TFIIH basal transcription factor comp... 32 0.87 sp|P46916|YTXO_BACSU Hypothetical protein ytxO (ORFY) 32 0.87
>sp|P32773|TOA1_YEAST Transcription initiation factor IIA large chain (TFIIA 32 kDa subunit) Length = 286 Score = 37.7 bits (86), Expect = 0.021 Identities = 22/72 (30%), Positives = 43/72 (59%), Gaps = 2/72 (2%) Frame = +1 Query: 4 VNFLNMSTVENITPVTEEKNN--LKEVIEKKTVDSENVDNDEPETKKQRLSDLNDIKDSE 177 +N N++TVENI +E+K++ +E +EK + E ++ + + KK++ S L D++ Sbjct: 160 INNANITTVENIDDESEKKDDEEKEEDVEKTRKEKEQIEQVKLQAKKEKRSAL---LDTD 216 Query: 178 EKAQNGDDAEED 213 E DD+++D Sbjct: 217 EVGSELDDSDDD 228
>sp|Q9N4M4|ANC1_CAEEL Nuclear anchorage protein 1 (Anchorage 1 protein) (Nesprin homolog) Length = 8545 Score = 37.7 bits (86), Expect = 0.021 Identities = 23/67 (34%), Positives = 36/67 (53%), Gaps = 2/67 (2%) Frame = +1 Query: 16 NMSTVENITPVTEEKNNLKEVIEKKTVDSENVDNDEPETKK--QRLSDLNDIKDSEEKAQ 189 N T+E +TP+ EE L + IEKK + V ++P + Q+L D+ I + EE Sbjct: 2043 NPETIETVTPILEEYTTLVDNIEKKLANEIEVPTNDPSRQDLVQQLQDV--ILECEEVVV 2100 Query: 190 NGDDAEE 210 N D+ E+ Sbjct: 2101 NCDNIEK 2107
>sp|P25980|UBF1B_XENLA Nucleolar transcription factor 1-B (Upstream binding factor 1-B) (UBF-1-B) (xUBF-2) Length = 701 Score = 35.8 bits (81), Expect = 0.079 Identities = 25/119 (21%), Positives = 54/119 (45%) Frame = +1 Query: 16 NMSTVENITPVTEEKNNLKEVIEKKTVDSENVDNDEPETKKQRLSDLNDIKDSEEKAQNG 195 N + ++ T + + K VI+ K+ D E+ ++DE E D +D +D E+ +++G Sbjct: 589 NTNKRKSTTKIQAPSSKSKLVIQSKSDDDEDDEDDEDEED----DDDDDDEDKEDSSEDG 644 Query: 196 DDAEEDQXXXXXXXXXXXXXXXXXXXSEKGTEEHVKEKDCNGSSAKVTENGDSAENEVN 372 D ++ E E+ + + +GSS+ + + DS++++ N Sbjct: 645 DSSDSSSDEDSEEGEENEDEEDEEDDDEDNEED--DDDNESGSSSSSSSSADSSDSDSN 701
>sp|P36044|MNN4_YEAST Protein MNN4 Length = 1178 Score = 33.9 bits (76), Expect = 0.30 Identities = 24/105 (22%), Positives = 40/105 (38%) Frame = +1 Query: 52 EEKNNLKEVIEKKTVDSENVDNDEPETKKQRLSDLNDIKDSEEKAQNGDDAEEDQXXXXX 231 EE+ KE EKK + E E E KK++ + K+ EEK + ++ ++ + Sbjct: 1047 EEEKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKQEEEEKKKKEEEEK 1106 Query: 232 XXXXXXXXXXXXXXSEKGTEEHVKEKDCNGSSAKVTENGDSAENE 366 K E+ K+K+ K E E+E Sbjct: 1107 KKQEEGEKMKNEDEENKKNEDEEKKKNEEEEKKKQEEKNKKNEDE 1151
Score = 30.0 bits (66), Expect = 4.3
Identities = 25/99 (25%), Positives = 41/99 (41%)
Frame = +1
Query: 70 KEVIEKKTVDSENVDNDEPETKKQRLSDLNDIKDSEEKAQNGDDAEEDQXXXXXXXXXXX 249
KE EKK + E E E KK++ + K+ EEK + ++ ++ Q
Sbjct: 1045 KEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKQ----------- 1093
Query: 250 XXXXXXXXSEKGTEEHVKEKDCNGSSAKVTENGDSAENE 366
+K EE K+K G K E+ ++ +NE
Sbjct: 1094 -----EEEEKKKKEEEEKKKQEEGEKMK-NEDEENKKNE 1126
Score = 29.3 bits (64), Expect = 7.4
Identities = 16/46 (34%), Positives = 26/46 (56%)
Frame = +1
Query: 52 EEKNNLKEVIEKKTVDSENVDNDEPETKKQRLSDLNDIKDSEEKAQ 189
E+K N +E EKK + +N N++ E KKQ + ++ E+K Q
Sbjct: 1129 EKKKNEEE--EKKKQEEKNKKNEDEEKKKQEEEEKKKNEEEEKKKQ 1172
Score = 28.9 bits (63), Expect = 9.6
Identities = 16/54 (29%), Positives = 29/54 (53%)
Frame = +1
Query: 55 EKNNLKEVIEKKTVDSENVDNDEPETKKQRLSDLNDIKDSEEKAQNGDDAEEDQ 216
EK ++ KK D E N+E E KKQ + + +D E+K Q ++ ++++
Sbjct: 1113 EKMKNEDEENKKNEDEEKKKNEEEEKKKQEEKNKKN-EDEEKKKQEEEEKKKNE 1165
>sp|P24788|CD2L1_MOUSE PITSLRE serine/threonine-protein kinase CDC2L1 (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) Length = 784 Score = 33.5 bits (75), Expect = 0.39 Identities = 22/90 (24%), Positives = 39/90 (43%) Frame = +1 Query: 97 DSENVDNDEPETKKQRLSDLNDIKDSEEKAQNGDDAEEDQXXXXXXXXXXXXXXXXXXXS 276 D+ +E ++ LSDL DI DSE K + + + + S Sbjct: 247 DNRKPVKEEKVEERDLLSDLQDISDSERKTSSAESSSAES----------------GSGS 290 Query: 277 EKGTEEHVKEKDCNGSSAKVTENGDSAENE 366 E+ EE +E++ GS+++ +E + E E Sbjct: 291 EEEEEEEEEEEEEEGSTSEESEEEEEEEEE 320
>sp|Q03101|CYAG_DICDI Adenylate cyclase, germination specific (ATP pyrophosphate-lyase) (Adenylyl cyclase) Length = 858 Score = 33.5 bits (75), Expect = 0.39 Identities = 21/76 (27%), Positives = 35/76 (46%), Gaps = 11/76 (14%) Frame = +1 Query: 7 NFLNMSTVENITPVTEEKNNLKEVIEKKTVDSENV-DNDEPETKKQRLSD---------- 153 N +N++ +N+ E NN + IE D+ N+ DN+ T + + Sbjct: 760 NDININNSDNVNNY-ENNNNFSDKIENNDGDNNNINDNNYKSTNENNIKSKTLFKDSKSL 818 Query: 154 LNDIKDSEEKAQNGDD 201 +NDIK ++E N DD Sbjct: 819 INDIKMAKENCDNNDD 834
>sp|Q6FSW2|RAD52_CANGA DNA repair and recombination protein RAD52 Length = 505 Score = 33.1 bits (74), Expect = 0.51 Identities = 13/32 (40%), Positives = 20/32 (62%) Frame = +1 Query: 100 SENVDNDEPETKKQRLSDLNDIKDSEEKAQNG 195 S +DND P K+Q+L++ N I D+ + NG Sbjct: 181 SNTIDNDNPAVKRQKLNNQNSIPDTNKVKYNG 212
>sp|Q6DRL5|MBB1A_BRARE Myb-binding protein 1A-like protein Length = 1269 Score = 32.7 bits (73), Expect = 0.67 Identities = 17/55 (30%), Positives = 28/55 (50%) Frame = +1 Query: 52 EEKNNLKEVIEKKTVDSENVDNDEPETKKQRLSDLNDIKDSEEKAQNGDDAEEDQ 216 ++K LKE E + E+ DNDE + + ++ EE + + DD EED+ Sbjct: 723 DKKRKLKEEDEDDDDEEEDDDNDEGDDDDDDDDEEEGGEEGEESSDSSDDEEEDE 777
>sp|Q960E8|TF2H1_DROME TFIIH basal transcription factor complex subunit 1 Length = 585 Score = 32.3 bits (72), Expect = 0.87 Identities = 15/35 (42%), Positives = 21/35 (60%) Frame = +1 Query: 100 SENVDNDEPETKKQRLSDLNDIKDSEEKAQNGDDA 204 S+ VD DEP++KKQRL + D + GDD+ Sbjct: 361 SDQVDKDEPQSKKQRLMEKIHYVDLGDPILEGDDS 395
>sp|P46916|YTXO_BACSU Hypothetical protein ytxO (ORFY) Length = 143 Score = 32.3 bits (72), Expect = 0.87 Identities = 21/72 (29%), Positives = 38/72 (52%) Frame = +1 Query: 1 EVNFLNMSTVENITPVTEEKNNLKEVIEKKTVDSENVDNDEPETKKQRLSDLNDIKDSEE 180 E L++ ++ + + K KE EK+T D + VD DE ETK + + + +DS+ Sbjct: 41 EEKHLDVGSIRELPHINAPKEYQKETKEKET-DRKIVD-DEEETKFETTLEPEEERDSKR 98 Query: 181 KAQNGDDAEEDQ 216 A ++ +ED+ Sbjct: 99 SAPPYEEEDEDE 110
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.316 0.133 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,599,100
Number of Sequences: 369166
Number of extensions: 625733
Number of successful extensions: 3961
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 2845
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3778
length of database: 68,354,980
effective HSP length: 105
effective length of database: 48,957,805
effective search space used: 4259329035
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
Cluster detail