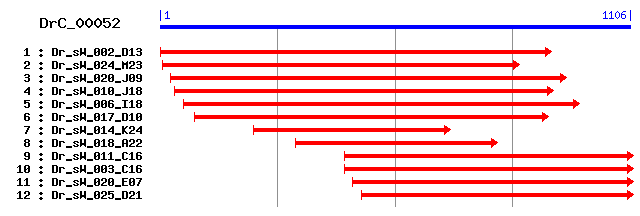
DrC_00052
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00052 (1089 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P60891|PRPS1_HUMAN Ribose-phosphate pyrophosphokinase I ... 325 1e-88 sp|Q9D7G0|PRPS1_MOUSE Ribose-phosphate pyrophosphokinase I ... 325 2e-88 sp|Q4R4R7|PRPS2_MACFA Ribose-phosphate pyrophosphokinase II... 323 5e-88 sp|P09330|PRPS2_RAT Ribose-phosphate pyrophosphokinase II (... 323 5e-88 sp|Q9CS42|PRPS2_MOUSE Ribose-phosphate pyrophosphokinase II... 323 7e-88 sp|P21108|PRPS3_HUMAN Ribose-phosphate pyrophosphokinase II... 317 3e-86 sp|P38689|KPR3_YEAST Ribose-phosphate pyrophosphokinase 3 (... 259 7e-69 sp|P38063|KPR4_YEAST Probable ribose-phosphate pyrophosphok... 258 2e-68 sp|P38620|KPR2_YEAST Ribose-phosphate pyrophosphokinase 2 (... 255 1e-67 sp|P46585|KPR1_CANAL Ribose-phosphate pyrophosphokinase 1 (... 254 4e-67
>sp|P60891|PRPS1_HUMAN Ribose-phosphate pyrophosphokinase I (Phosphoribosyl pyrophosphate synthetase I) (PRS-I) (PPRibP) sp|P60892|PRPS1_RAT Ribose-phosphate pyrophosphokinase I (Phosphoribosyl pyrophosphate synthetase I) (PRS-I) Length = 318 Score = 325 bits (833), Expect = 1e-88 Identities = 163/316 (51%), Positives = 235/316 (74%), Gaps = 2/316 (0%) Frame = +3 Query: 24 MLNIRLFSGSSNILLTEKIGERIGIKLSEAILNKFSNNETSVQIKESVRGKDVFILQSGY 203 M NI++FSGSS+ L++KI +R+G++L + + KFSN ET V+I ESVRG+DV+I+QSG Sbjct: 1 MPNIKIFSGSSHQDLSQKIADRLGLELGKVVTKKFSNQETCVEIGESVRGEDVYIVQSGC 60 Query: 204 IDVNNHLMELLIMINACKNASSARITAVLPYMPYSRQSKKDNNRVPITAKLVADLLHIAG 383 ++N++LMELLIMINACK AS++R+TAV+P PY+RQ KKD +R PI+AKLVA++L +AG Sbjct: 61 GEINDNLMELLIMINACKIASASRVTAVIPCFPYARQDKKDKSRAPISAKLVANMLSVAG 120 Query: 384 ANHIITMDLHASQIQGFFNVPVDNLLSESCHISWIKKNISKWEEITIFSLNVTSAERVAN 563 A+HIITMDLHASQIQGFF++PVDNL +E + WI++NIS+W TI S + A+RV + Sbjct: 121 ADHIITMDLHASQIQGFFDIPVDNLYAEPAVLKWIRENISEWRNCTIVSPDAGGAKRVTS 180 Query: 564 MANTLKVRFGLIYVEKKEASE--QIILVGDVVSRIAVILLDIIDNCDLVISSVKRLKEVG 737 +A+ L V F LI+ E+K+A+E +++LVGDV R+A+++ D+ D C + + +L G Sbjct: 181 IADRLNVDFALIHKERKKANEVDRMVLVGDVKDRVAILVDDMADTCGTICHAADKLLSAG 240 Query: 738 AAGVYVIATHGMLSERLMKEVDDSIITKLVVTNTICQEKYVKQSDKIACLDVSVIFAEAI 917 A VY I THG+ S + ++++ +VVTNTI QE +K KI +D+S+I AEAI Sbjct: 241 ATRVYAILTHGIFSGPAISRINNACFEAVVVTNTIPQEDKMKHCSKIQVIDISMILAEAI 300 Query: 918 RRIHYGESVSCLYNNL 965 RR H GESVS L++++ Sbjct: 301 RRTHNGESVSYLFSHV 316
>sp|Q9D7G0|PRPS1_MOUSE Ribose-phosphate pyrophosphokinase I (Phosphoribosyl pyrophosphate synthetase I) (PRS-I) Length = 318 Score = 325 bits (832), Expect = 2e-88 Identities = 163/316 (51%), Positives = 234/316 (74%), Gaps = 2/316 (0%) Frame = +3 Query: 24 MLNIRLFSGSSNILLTEKIGERIGIKLSEAILNKFSNNETSVQIKESVRGKDVFILQSGY 203 M NI++FSGSS+ L++KI +R+G++L + + KFSN ET V+I ESVRG+DV+I+QSG Sbjct: 1 MPNIKIFSGSSHQDLSQKIADRLGLELGKVVTKKFSNQETCVEIGESVRGEDVYIVQSGC 60 Query: 204 IDVNNHLMELLIMINACKNASSARITAVLPYMPYSRQSKKDNNRVPITAKLVADLLHIAG 383 ++N++LMELLIMINACK AS++R+TAV+P PY+RQ KKD +R PI+AKLVA++L +AG Sbjct: 61 GEINDNLMELLIMINACKIASASRVTAVIPCFPYARQDKKDKSRAPISAKLVANMLSVAG 120 Query: 384 ANHIITMDLHASQIQGFFNVPVDNLLSESCHISWIKKNISKWEEITIFSLNVTSAERVAN 563 A+HIITMDLHASQIQGFF++PVDNL +E + WI++NIS+W TI S + A+RV + Sbjct: 121 ADHIITMDLHASQIQGFFDIPVDNLYAEPAVLKWIRENISEWRNCTIVSPDAGGAKRVTS 180 Query: 564 MANTLKVRFGLIYVEKKEASE--QIILVGDVVSRIAVILLDIIDNCDLVISSVKRLKEVG 737 +A L V F LI+ E+K+A+E +++LVGDV R+A+++ D+ D C + + +L G Sbjct: 181 IAERLNVDFALIHKERKKANEVDRMVLVGDVKDRVAILVDDMADTCGTICHAADKLLSAG 240 Query: 738 AAGVYVIATHGMLSERLMKEVDDSIITKLVVTNTICQEKYVKQSDKIACLDVSVIFAEAI 917 A VY I THG+ S + ++++ +VVTNTI QE +K KI +D+S+I AEAI Sbjct: 241 ATRVYAILTHGIFSGPAISRINNACFEAVVVTNTIPQEDKMKHCSKIQVIDISMILAEAI 300 Query: 918 RRIHYGESVSCLYNNL 965 RR H GESVS L++++ Sbjct: 301 RRTHNGESVSYLFSHV 316
>sp|Q4R4R7|PRPS2_MACFA Ribose-phosphate pyrophosphokinase II (Phosphoribosyl pyrophosphate synthetase II) (PRS-II) sp|P11908|PRPS2_HUMAN Ribose-phosphate pyrophosphokinase II (Phosphoribosyl pyrophosphate synthetase II) (PRS-II) (PPRibP) Length = 318 Score = 323 bits (828), Expect = 5e-88 Identities = 162/316 (51%), Positives = 235/316 (74%), Gaps = 2/316 (0%) Frame = +3 Query: 24 MLNIRLFSGSSNILLTEKIGERIGIKLSEAILNKFSNNETSVQIKESVRGKDVFILQSGY 203 M NI LFSGSS+ L++++ +R+G++L + + KFSN ETSV+I ESVRG+DV+I+QSG Sbjct: 1 MPNIVLFSGSSHQDLSQRVADRLGLELGKVVTKKFSNQETSVEIGESVRGEDVYIIQSGC 60 Query: 204 IDVNNHLMELLIMINACKNASSARITAVLPYMPYSRQSKKDNNRVPITAKLVADLLHIAG 383 ++N++LMELLIMINACK ASS+R+TAV+P PY+RQ KKD +R PI+AKLVA++L +AG Sbjct: 61 GEINDNLMELLIMINACKIASSSRVTAVIPCFPYARQDKKDKSRAPISAKLVANMLSVAG 120 Query: 384 ANHIITMDLHASQIQGFFNVPVDNLLSESCHISWIKKNISKWEEITIFSLNVTSAERVAN 563 A+HIITMDLHASQIQGFF++PVDNL +E + WI++NI++W+ I S + A+RV + Sbjct: 121 ADHIITMDLHASQIQGFFDIPVDNLYAEPAVLQWIRENIAEWKNCIIVSPDAGGAKRVTS 180 Query: 564 MANTLKVRFGLIYVEKKEASE--QIILVGDVVSRIAVILLDIIDNCDLVISSVKRLKEVG 737 +A+ L V F LI+ E+K+A+E +++LVGDV R+A+++ D+ D C + + +L G Sbjct: 181 IADRLNVEFALIHKERKKANEVDRMVLVGDVKDRVAILVDDMADTCGTICHAADKLLSAG 240 Query: 738 AAGVYVIATHGMLSERLMKEVDDSIITKLVVTNTICQEKYVKQSDKIACLDVSVIFAEAI 917 A VY I THG+ S + ++++ +VVTNTI QE +K KI +D+S+I AEAI Sbjct: 241 ATKVYAILTHGIFSGPAISRINNAAFEAVVVTNTIPQEDKMKHCTKIQVIDISMILAEAI 300 Query: 918 RRIHYGESVSCLYNNL 965 RR H GESVS L++++ Sbjct: 301 RRTHNGESVSYLFSHV 316
>sp|P09330|PRPS2_RAT Ribose-phosphate pyrophosphokinase II (Phosphoribosyl pyrophosphate synthetase II) (PRS-II) Length = 318 Score = 323 bits (828), Expect = 5e-88 Identities = 162/316 (51%), Positives = 234/316 (74%), Gaps = 2/316 (0%) Frame = +3 Query: 24 MLNIRLFSGSSNILLTEKIGERIGIKLSEAILNKFSNNETSVQIKESVRGKDVFILQSGY 203 M NI LFSGSS+ L++++ +R+G++L + + KFSN ETSV+I ESVRG+DV+I+QSG Sbjct: 1 MPNIVLFSGSSHQDLSQRVADRLGLELGKVVTKKFSNQETSVEIGESVRGEDVYIIQSGC 60 Query: 204 IDVNNHLMELLIMINACKNASSARITAVLPYMPYSRQSKKDNNRVPITAKLVADLLHIAG 383 ++N++LMELLIMINACK ASS+R+TAV+P PY+RQ KKD +R PI+AKLVA++L +AG Sbjct: 61 GEINDNLMELLIMINACKIASSSRVTAVIPCFPYARQDKKDKSRAPISAKLVANMLSVAG 120 Query: 384 ANHIITMDLHASQIQGFFNVPVDNLLSESCHISWIKKNISKWEEITIFSLNVTSAERVAN 563 A+HIITMDLHASQIQGFF++PVDNL +E + WI++NI++W I S + A+RV + Sbjct: 121 ADHIITMDLHASQIQGFFDIPVDNLYAEPAVLQWIRENITEWRNCIIVSPDAGGAKRVTS 180 Query: 564 MANTLKVRFGLIYVEKKEASE--QIILVGDVVSRIAVILLDIIDNCDLVISSVKRLKEVG 737 +A+ L V F LI+ E+K+A+E +++LVGDV R+A+++ D+ D C + + +L G Sbjct: 181 IADRLNVEFALIHKERKKANEVDRMVLVGDVKDRVAILVDDMADTCGTICHAADKLLSAG 240 Query: 738 AAGVYVIATHGMLSERLMKEVDDSIITKLVVTNTICQEKYVKQSDKIACLDVSVIFAEAI 917 A VY I THG+ S + ++++ +VVTNTI QE +K KI +D+S+I AEAI Sbjct: 241 ATKVYAILTHGIFSGPAISRINNAAFEAVVVTNTIPQEDKMKHCSKIQVIDISMILAEAI 300 Query: 918 RRIHYGESVSCLYNNL 965 RR H GESVS L++++ Sbjct: 301 RRTHNGESVSYLFSHV 316
>sp|Q9CS42|PRPS2_MOUSE Ribose-phosphate pyrophosphokinase II (Phosphoribosyl pyrophosphate synthetase II) (PRS-II) Length = 318 Score = 323 bits (827), Expect = 7e-88 Identities = 162/316 (51%), Positives = 233/316 (73%), Gaps = 2/316 (0%) Frame = +3 Query: 24 MLNIRLFSGSSNILLTEKIGERIGIKLSEAILNKFSNNETSVQIKESVRGKDVFILQSGY 203 M NI LFSGSS+ L++++ +R+G++L + + KFSN ETSV+I ESVRG+DV+I+QSG Sbjct: 1 MPNIVLFSGSSHQDLSQRVADRLGLELGKVVTKKFSNQETSVEIGESVRGEDVYIIQSGC 60 Query: 204 IDVNNHLMELLIMINACKNASSARITAVLPYMPYSRQSKKDNNRVPITAKLVADLLHIAG 383 ++N++LMELLIMINACK ASS+R+TAV+P PY+RQ KKD +R PI+AKLVA++L +AG Sbjct: 61 GEINDNLMELLIMINACKIASSSRVTAVIPCFPYARQDKKDKSRAPISAKLVANMLSVAG 120 Query: 384 ANHIITMDLHASQIQGFFNVPVDNLLSESCHISWIKKNISKWEEITIFSLNVTSAERVAN 563 A+HIITMDLHASQIQGFF++PVDNL +E + WI++NI++W I S + A+RV + Sbjct: 121 ADHIITMDLHASQIQGFFDIPVDNLYAEPAVLQWIRENITEWRNCIIVSPDAGGAKRVTS 180 Query: 564 MANTLKVRFGLIYVEKKEASE--QIILVGDVVSRIAVILLDIIDNCDLVISSVKRLKEVG 737 +A+ L V F LI+ E+K+A+E +++LVGDV R+A+++ D+ D C + + +L G Sbjct: 181 IADRLNVEFALIHKERKKANEVDRMVLVGDVKDRVAILVDDMADTCGTICHAADKLLSAG 240 Query: 738 AAGVYVIATHGMLSERLMKEVDDSIITKLVVTNTICQEKYVKQSDKIACLDVSVIFAEAI 917 A VY I THG+ S + ++ + +VVTNTI QE +K KI +D+S+I AEAI Sbjct: 241 ATKVYAILTHGIFSGPAISRINSAAFEAVVVTNTIPQEDKMKHCSKIQVIDISMILAEAI 300 Query: 918 RRIHYGESVSCLYNNL 965 RR H GESVS L++++ Sbjct: 301 RRTHNGESVSYLFSHV 316
>sp|P21108|PRPS3_HUMAN Ribose-phosphate pyrophosphokinase III (Phosphoribosyl pyrophosphate synthetase III) (PRS-III) (Phosphoribosyl pyrophosphate synthetase 1-like 1) Length = 318 Score = 317 bits (813), Expect = 3e-86 Identities = 162/316 (51%), Positives = 232/316 (73%), Gaps = 2/316 (0%) Frame = +3 Query: 24 MLNIRLFSGSSNILLTEKIGERIGIKLSEAILNKFSNNETSVQIKESVRGKDVFILQSGY 203 M NI++FSGSS+ L++KI +R+G++L + + KFSN ET V+I ESVRG+DV+I+QSG Sbjct: 1 MPNIKIFSGSSHQDLSQKIADRLGLELGKVVTKKFSNQETCVEIDESVRGEDVYIVQSGC 60 Query: 204 IDVNNHLMELLIMINACKNASSARITAVLPYMPYSRQSKKDNNRVPITAKLVADLLHIAG 383 ++N+ LMELLIMINACK AS++R+TAV+P PY+RQ KKD +R PI+AKLVA++L IAG Sbjct: 61 GEINDSLMELLIMINACKIASASRVTAVIPCFPYARQDKKDKSRSPISAKLVANMLSIAG 120 Query: 384 ANHIITMDLHASQIQGFFNVPVDNLLSESCHISWIKKNISKWEEITIFSLNVTSAERVAN 563 A+HIITMDLHASQIQGFF++PVDNL +E + WI++NI +W+ I S + A+RV + Sbjct: 121 ADHIITMDLHASQIQGFFDIPVDNLYAEPTVLKWIRENIPEWKNCIIVSPDAGGAKRVTS 180 Query: 564 MANTLKVRFGLIYVEKKEASEQ--IILVGDVVSRIAVILLDIIDNCDLVISSVKRLKEVG 737 +A+ L V F LI+ E+K+A+E I+LVGDV R+A+++ D+ D C + + +L G Sbjct: 181 IADQLNVDFALIHKERKKANEVDCIVLVGDVNDRVAILVDDMADTCVTICLAADKLLSAG 240 Query: 738 AAGVYVIATHGMLSERLMKEVDDSIITKLVVTNTICQEKYVKQSDKIACLDVSVIFAEAI 917 A VY I THG+ S + ++ + +VVTNTI Q++ +K KI +D+S+I AEAI Sbjct: 241 ATRVYAILTHGIFSGPAISRINTACFEAVVVTNTIPQDEKMKHCSKIRVIDISMILAEAI 300 Query: 918 RRIHYGESVSCLYNNL 965 RR H GESVS L++++ Sbjct: 301 RRTHNGESVSYLFSHV 316
>sp|P38689|KPR3_YEAST Ribose-phosphate pyrophosphokinase 3 (Phosphoribosyl pyrophosphate synthetase 3) Length = 320 Score = 259 bits (663), Expect = 7e-69 Identities = 139/314 (44%), Positives = 214/314 (68%), Gaps = 3/314 (0%) Frame = +3 Query: 30 NIRLFSGSSNILLTEKIGERIGIKLSEAILNKFSNNETSVQIKESVRGKDVFIL-QSGYI 206 +I+L + + L E + +R+G++L+ + L + E S I ESVR +D+FI+ Q G Sbjct: 5 SIKLLAPDVHRGLAELVAKRLGLQLTSSKLKRDPTGEVSFSIGESVRDQDIFIITQIGSG 64 Query: 207 DVNNHLMELLIMINACKNASSARITAVLPYMPYSRQSKKDNNRVPITAKLVADLLHIAGA 386 VN+ ++ELLIMINA K AS+ RITA++P PY+RQ +KD +R PITAKL+AD+L AG Sbjct: 65 VVNDRVLELLIMINASKTASARRITAIIPNFPYARQDRKDKSRAPITAKLMADMLTTAGC 124 Query: 387 NHIITMDLHASQIQGFFNVPVDNLLSESCHISWIKKNISKWEEITIFSLNVTSAERVANM 566 +H+ITMDLHASQIQGFF+VPVDNL +E + +IK+N++ + I I S + A+R A + Sbjct: 125 DHVITMDLHASQIQGFFDVPVDNLYAEPSVVRYIKENVNYMDSI-IISPDAGGAKRAATL 183 Query: 567 ANTLKVRFGLIYVEKKEASE--QIILVGDVVSRIAVILLDIIDNCDLVISSVKRLKEVGA 740 A+ L + F LI+ E+ A+E +++LVGDV +I +I+ D+ D C + + + L E A Sbjct: 184 ADRLDLNFALIHKERARANEVSRMVLVGDVTDKICIIVDDMADTCGTLAKAAEILLENRA 243 Query: 741 AGVYVIATHGMLSERLMKEVDDSIITKLVVTNTICQEKYVKQSDKIACLDVSVIFAEAIR 920 V I THG+LS R ++ +++S + ++V TNT+ E+ +K+ K+A +D+S + AE+IR Sbjct: 244 KSVIAIVTHGVLSGRAIENINNSKLDRVVCTNTVPFEEKIKKCPKLAVIDISSVLAESIR 303 Query: 921 RIHYGESVSCLYNN 962 R+H GES+S L+ N Sbjct: 304 RLHNGESISYLFKN 317
>sp|P38063|KPR4_YEAST Probable ribose-phosphate pyrophosphokinase 4 (Phosphoribosyl pyrophosphate synthetase 4) Length = 355 Score = 258 bits (660), Expect = 2e-68 Identities = 139/315 (44%), Positives = 222/315 (70%), Gaps = 4/315 (1%) Frame = +3 Query: 30 NIRLFSGSSNILLTEKIGERIGIKLSEAILNKFSNNETSVQIKESVRGKDVFILQSGY-- 203 +I+L +G+S+ L E+I +++GI LS+ + ++SN ETSV I ES+R +DV+I+Q+G Sbjct: 41 SIKLLAGNSHPDLAEQISKKLGIPLSKVGVYQYSNKETSVTIGESLRDEDVYIIQTGIGE 100 Query: 204 IDVNNHLMELLIMINACKNASSARITAVLPYMPYSRQSKKDNNRVPITAKLVADLLHIAG 383 ++N+ LMELLI+I+AC+ AS+ +IT V+P PY+RQ KKD +R PITAKLVA+LL AG Sbjct: 101 QEINDFLMELLILIHACQIASARKITTVIPNFPYARQDKKDKSRAPITAKLVANLLQTAG 160 Query: 384 ANHIITMDLHASQIQGFFNVPVDNLLSESCHISWIKKNISKWEEITIFSLNVTSAERVAN 563 A+H+ITMDLHASQIQGFF++PVDNL +E +++I+ + ++ + S + A+RVA Sbjct: 161 ADHVITMDLHASQIQGFFHIPVDNLYAEPSVLNYIRARKTDFDNAILVSPDAGGAKRVAA 220 Query: 564 MANTLKVRFGLIYVEKKEASE--QIILVGDVVSRIAVILLDIIDNCDLVISSVKRLKEVG 737 +A+ L + F LI+ E+++A+E +++LVGDV ++ +++ D+ D C ++ + L E G Sbjct: 221 LADKLDLNFALIHKERQKANEVSKMVLVGDVTNKSCLLVDDMADTCGTLVKACDTLMEHG 280 Query: 738 AAGVYVIATHGMLSERLMKEVDDSIITKLVVTNTICQEKYVKQSDKIACLDVSVIFAEAI 917 A V I THG+ S +++ +S ++++V TNT+ + + +D+I D+S FAEAI Sbjct: 281 AKEVIAIVTHGIFSGSAREKLRNSRLSRIVCTNTVPVDLDLPIADQI---DISPTFAEAI 337 Query: 918 RRIHYGESVSCLYNN 962 RR+H GESVS L+ + Sbjct: 338 RRLHNGESVSYLFTH 352
>sp|P38620|KPR2_YEAST Ribose-phosphate pyrophosphokinase 2 (Phosphoribosyl pyrophosphate synthetase 2) Length = 318 Score = 255 bits (652), Expect = 1e-67 Identities = 138/315 (43%), Positives = 217/315 (68%), Gaps = 4/315 (1%) Frame = +3 Query: 30 NIRLFSGSSNILLTEKIGERIGIKLSEAILNKFSNNETSVQIKESVRGKDVFILQSGY-- 203 +I+L +G+S+ L E I +R+G+ LS+ + ++SN ETSV I ES+R +DV+I+Q+GY Sbjct: 5 SIKLLAGNSHPGLAELISQRLGVPLSKVGVYQYSNKETSVTIGESIRDEDVYIIQTGYGE 64 Query: 204 IDVNNHLMELLIMINACKNASSARITAVLPYMPYSRQSKKDNNRVPITAKLVADLLHIAG 383 ++N+ LMELLI+I+ACK AS RITAV+P PY+RQ KKD +R PITAKL+A+LL AG Sbjct: 65 HEINDFLMELLILIHACKTASVRRITAVIPNFPYARQDKKDKSRAPITAKLIANLLETAG 124 Query: 384 ANHIITMDLHASQIQGFFNVPVDNLLSESCHISWIKKNISKWEEITIFSLNVTSAERVAN 563 +H+ITMDLHASQIQGFF++PVDNL E +++I+ + + + S + A+RVA+ Sbjct: 125 CDHVITMDLHASQIQGFFHIPVDNLYGEPSVLNYIRTK-TDFNNAILVSPDAGGAKRVAS 183 Query: 564 MANTLKVRFGLIYVEKKEASE--QIILVGDVVSRIAVILLDIIDNCDLVISSVKRLKEVG 737 +A+ L + F LI+ E+++A+E +++LVGDV + +++ D+ D C ++ + L + G Sbjct: 184 LADKLDMNFALIHKERQKANEVSRMLLVGDVAGKSCLLIDDMADTCGTLVKACDTLMDHG 243 Query: 738 AAGVYVIATHGMLSERLMKEVDDSIITKLVVTNTICQEKYVKQSDKIACLDVSVIFAEAI 917 A V I THG+ S +++ +S ++++V TNT+ + + D++ D+S AEAI Sbjct: 244 AKEVIAIVTHGIFSGSAREKLINSRLSRIVCTNTVPVDLDLDIVDQV---DISPTIAEAI 300 Query: 918 RRIHYGESVSCLYNN 962 RR+H GESVS L+ + Sbjct: 301 RRLHNGESVSYLFTH 315
>sp|P46585|KPR1_CANAL Ribose-phosphate pyrophosphokinase 1 (Phosphoribosyl pyrophosphate synthetase 1) Length = 321 Score = 254 bits (648), Expect = 4e-67 Identities = 138/314 (43%), Positives = 207/314 (65%), Gaps = 3/314 (0%) Frame = +3 Query: 30 NIRLFSGSSNILLTEKIGERIGIKLSEAILNKFSNNETSVQIKESVRGKDVFIL-QSGYI 206 +I+L + + L E + +R+G++L+ + L + S E I ESVR +D+FI+ Q G Sbjct: 5 SIKLLASDVHRGLAELVSKRLGLRLTPSELKRESTGEVQFSIGESVRDEDIFIICQIGSG 64 Query: 207 DVNNHLMELLIMINACKNASSARITAVLPYMPYSRQSKKDNNRVPITAKLVADLLHIAGA 386 +VN+ + EL+IMINACK AS+ RIT +LP PY+RQ +KD +R PITAKL+AD+L AG Sbjct: 65 EVNDRVFELMIMINACKTASARRITVILPNFPYARQDRKDKSRAPITAKLMADMLTTAGC 124 Query: 387 NHIITMDLHASQIQGFFNVPVDNLLSESCHISWIKKNISKWEEITIFSLNVTSAERVANM 566 +H+ITMDLHASQIQGFF+VPVDNL +E + +IK+ I + E I S + A+R A + Sbjct: 125 DHVITMDLHASQIQGFFDVPVDNLYAEPSVVRYIKETID-YSEAIIISSDAGGAKRAAGL 183 Query: 567 ANTLKVRFGLIYVEKKEASE--QIILVGDVVSRIAVILLDIIDNCDLVISSVKRLKEVGA 740 A+ L + F LI+ E+ A+E +++LV DV +I VI+ D+ D C + + + L + A Sbjct: 184 ADRLDLNFELIHKERARANEVSRMVLVVDVTDKICVIVDDMADTCGTLAKAAEVLLDNNA 243 Query: 741 AGVYVIATHGMLSERLMKEVDDSIITKLVVTNTICQEKYVKQSDKIACLDVSVIFAEAIR 920 V I THG+LS +K +++S + K+V TNT+ E +K K+ +D+S + AE+IR Sbjct: 244 KDVIAIVTHGILSGNAIKNINNSKLKKVVCTNTVPFEDKLKLCLKLDTIDISAVIAESIR 303 Query: 921 RIHYGESVSCLYNN 962 R+H GES+S L+ N Sbjct: 304 RLHNGESISYLFKN 317
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 121,202,982
Number of Sequences: 369166
Number of extensions: 2474120
Number of successful extensions: 6467
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5901
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6237
length of database: 68,354,980
effective HSP length: 112
effective length of database: 47,664,660
effective search space used: 11916165000
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail