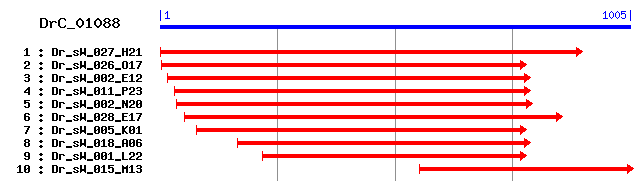
DrC_01088
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01088 (1005 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9CQD1|RAB5A_MOUSE Ras-related protein Rab-5A 262 1e-69 sp|P20339|RAB5A_HUMAN Ras-related protein Rab-5A 262 1e-69 sp|P18066|RAB5A_CANFA Ras-related protein Rab-5A >gi|471170... 262 1e-69 sp|P51148|RAB5C_HUMAN Ras-related protein Rab-5C (RAB5L) (L... 256 5e-68 sp|P51147|RAB5C_CANFA Ras-related protein Rab-5C 256 9e-68 sp|P35278|RAB5C_MOUSE Ras-related protein Rab-5C 256 9e-68 sp|P61021|RAB5B_MOUSE Ras-related protein Rab-5B >gi|465776... 256 9e-68 sp|P29687|RAB5_TOBAC Ras-related protein Rab5 228 3e-59 sp|P31582|RHA1_ARATH Ras-related protein RHA1 226 6e-59 sp|P31583|RHN1_NICPL Ras-related protein RHN1 226 8e-59
>sp|Q9CQD1|RAB5A_MOUSE Ras-related protein Rab-5A Length = 215 Score = 262 bits (669), Expect = 1e-69 Identities = 130/180 (72%), Positives = 153/180 (85%) Frame = +1 Query: 34 NRQVQFKLVLLGESAVGKSSLVMRFVKGQFHEYQESTVGAAFLTQTVVVEDTTVKFEIWD 213 N+ QFKLVLLGESAVGKSSLV+RFVKGQFHE+QEST+GAAFLTQTV ++DTTVKFEIWD Sbjct: 16 NKICQFKLVLLGESAVGKSSLVLRFVKGQFHEFQESTIGAAFLTQTVCLDDTTVKFEIWD 75 Query: 214 TAGQERYHSLAPMYYRGAQAAIVVYDITSQESFHRAKTWIQELTEKATSVKVIALAGNKS 393 TAGQERYHSLAPMYYRGAQAAIVVYDIT++ESF RAK W++EL +A+ VIAL+GNK+ Sbjct: 76 TAGQERYHSLAPMYYRGAQAAIVVYDITNEESFARAKNWVKELQRQASPNIVIALSGNKA 135 Query: 394 DLSAQREVSISDGSTYAQENGLLFMETSAKTSSNVNEIFLEIAKKLPRHEVTTPNAGSGQ 573 DL+ +R V + +YA +N LLFMETSAKTS NVNEIF+ IAKKLP++E P A S + Sbjct: 136 DLANKRAVDFQEAQSYADDNSLLFMETSAKTSMNVNEIFMAIAKKLPKNEPQNPGANSAR 195
>sp|P20339|RAB5A_HUMAN Ras-related protein Rab-5A Length = 215 Score = 262 bits (669), Expect = 1e-69 Identities = 130/180 (72%), Positives = 153/180 (85%) Frame = +1 Query: 34 NRQVQFKLVLLGESAVGKSSLVMRFVKGQFHEYQESTVGAAFLTQTVVVEDTTVKFEIWD 213 N+ QFKLVLLGESAVGKSSLV+RFVKGQFHE+QEST+GAAFLTQTV ++DTTVKFEIWD Sbjct: 16 NKICQFKLVLLGESAVGKSSLVLRFVKGQFHEFQESTIGAAFLTQTVCLDDTTVKFEIWD 75 Query: 214 TAGQERYHSLAPMYYRGAQAAIVVYDITSQESFHRAKTWIQELTEKATSVKVIALAGNKS 393 TAGQERYHSLAPMYYRGAQAAIVVYDIT++ESF RAK W++EL +A+ VIAL+GNK+ Sbjct: 76 TAGQERYHSLAPMYYRGAQAAIVVYDITNEESFARAKNWVKELQRQASPNIVIALSGNKA 135 Query: 394 DLSAQREVSISDGSTYAQENGLLFMETSAKTSSNVNEIFLEIAKKLPRHEVTTPNAGSGQ 573 DL+ +R V + +YA +N LLFMETSAKTS NVNEIF+ IAKKLP++E P A S + Sbjct: 136 DLANKRAVDFQEAQSYADDNSLLFMETSAKTSMNVNEIFMAIAKKLPKNEPQNPGANSAR 195
>sp|P18066|RAB5A_CANFA Ras-related protein Rab-5A sp|P61271|RAB5A_MACFA Ras-related protein Rab-5A Length = 215 Score = 262 bits (669), Expect = 1e-69 Identities = 130/180 (72%), Positives = 153/180 (85%) Frame = +1 Query: 34 NRQVQFKLVLLGESAVGKSSLVMRFVKGQFHEYQESTVGAAFLTQTVVVEDTTVKFEIWD 213 N+ QFKLVLLGESAVGKSSLV+RFVKGQFHE+QEST+GAAFLTQTV ++DTTVKFEIWD Sbjct: 16 NKICQFKLVLLGESAVGKSSLVLRFVKGQFHEFQESTIGAAFLTQTVCLDDTTVKFEIWD 75 Query: 214 TAGQERYHSLAPMYYRGAQAAIVVYDITSQESFHRAKTWIQELTEKATSVKVIALAGNKS 393 TAGQERYHSLAPMYYRGAQAAIVVYDIT++ESF RAK W++EL +A+ VIAL+GNK+ Sbjct: 76 TAGQERYHSLAPMYYRGAQAAIVVYDITNEESFARAKNWVKELQRQASPNIVIALSGNKA 135 Query: 394 DLSAQREVSISDGSTYAQENGLLFMETSAKTSSNVNEIFLEIAKKLPRHEVTTPNAGSGQ 573 DL+ +R V + +YA +N LLFMETSAKTS NVNEIF+ IAKKLP++E P A S + Sbjct: 136 DLANKRAVDFQEAQSYADDNSLLFMETSAKTSMNVNEIFMAIAKKLPKNEPQNPGANSAR 195
>sp|P51148|RAB5C_HUMAN Ras-related protein Rab-5C (RAB5L) (L1880) Length = 216 Score = 256 bits (655), Expect = 5e-68 Identities = 127/180 (70%), Positives = 150/180 (83%) Frame = +1 Query: 34 NRQVQFKLVLLGESAVGKSSLVMRFVKGQFHEYQESTVGAAFLTQTVVVEDTTVKFEIWD 213 N+ QFKLVLLGESAVGKSSLV+RFVKGQFHEYQEST+GAAFLTQTV ++DTTVKFEIWD Sbjct: 17 NKICQFKLVLLGESAVGKSSLVLRFVKGQFHEYQESTIGAAFLTQTVCLDDTTVKFEIWD 76 Query: 214 TAGQERYHSLAPMYYRGAQAAIVVYDITSQESFHRAKTWIQELTEKATSVKVIALAGNKS 393 TAGQERYHSLAPMYYRGAQAAIVVYDIT+ ++F RAK W++EL +A+ VIALAGNK+ Sbjct: 77 TAGQERYHSLAPMYYRGAQAAIVVYDITNTDTFARAKNWVKELQRQASPNIVIALAGNKA 136 Query: 394 DLSAQREVSISDGSTYAQENGLLFMETSAKTSSNVNEIFLEIAKKLPRHEVTTPNAGSGQ 573 DL+++R V + YA +N LLFMETSAKT+ NVNEIF+ IAKKLP++E G+ Sbjct: 137 DLASKRAVEFQEAQAYADDNSLLFMETSAKTAMNVNEIFMAIAKKLPKNEPQNATGAPGR 196
>sp|P51147|RAB5C_CANFA Ras-related protein Rab-5C Length = 216 Score = 256 bits (653), Expect = 9e-68 Identities = 126/170 (74%), Positives = 148/170 (87%) Frame = +1 Query: 34 NRQVQFKLVLLGESAVGKSSLVMRFVKGQFHEYQESTVGAAFLTQTVVVEDTTVKFEIWD 213 N+ QFKLVLLGESAVGKSSLV+RFVKGQFHEYQEST+GAAFLTQTV ++DTTVKFEIWD Sbjct: 17 NKICQFKLVLLGESAVGKSSLVLRFVKGQFHEYQESTIGAAFLTQTVCLDDTTVKFEIWD 76 Query: 214 TAGQERYHSLAPMYYRGAQAAIVVYDITSQESFHRAKTWIQELTEKATSVKVIALAGNKS 393 TAGQERYHSLAPMYYRGAQAAIVVYDIT+ ++F RAK W++EL +A+ VIALAGNK+ Sbjct: 77 TAGQERYHSLAPMYYRGAQAAIVVYDITNTDTFARAKNWVKELQRQASPNIVIALAGNKA 136 Query: 394 DLSAQREVSISDGSTYAQENGLLFMETSAKTSSNVNEIFLEIAKKLPRHE 543 DL+++R V + YA +N LLFMETSAKT+ NVNEIF+ IAKKLP++E Sbjct: 137 DLASKRAVEFQEAQAYADDNSLLFMETSAKTAMNVNEIFMAIAKKLPKNE 186
>sp|P35278|RAB5C_MOUSE Ras-related protein Rab-5C Length = 216 Score = 256 bits (653), Expect = 9e-68 Identities = 126/170 (74%), Positives = 148/170 (87%) Frame = +1 Query: 34 NRQVQFKLVLLGESAVGKSSLVMRFVKGQFHEYQESTVGAAFLTQTVVVEDTTVKFEIWD 213 N+ QFKLVLLGESAVGKSSLV+RFVKGQFHEYQEST+GAAFLTQTV ++DTTVKFEIWD Sbjct: 17 NKICQFKLVLLGESAVGKSSLVLRFVKGQFHEYQESTIGAAFLTQTVCLDDTTVKFEIWD 76 Query: 214 TAGQERYHSLAPMYYRGAQAAIVVYDITSQESFHRAKTWIQELTEKATSVKVIALAGNKS 393 TAGQERYHSLAPMYYRGAQAAIVVYDIT+ ++F RAK W++EL +A+ VIALAGNK+ Sbjct: 77 TAGQERYHSLAPMYYRGAQAAIVVYDITNTDTFARAKNWVKELQRQASPNIVIALAGNKA 136 Query: 394 DLSAQREVSISDGSTYAQENGLLFMETSAKTSSNVNEIFLEIAKKLPRHE 543 DL+++R V + YA +N LLFMETSAKT+ NVNEIF+ IAKKLP++E Sbjct: 137 DLASKRAVEFQEAQAYADDNSLLFMETSAKTAMNVNEIFMAIAKKLPKNE 186
>sp|P61021|RAB5B_MOUSE Ras-related protein Rab-5B sp|P61020|RAB5B_HUMAN Ras-related protein Rab-5B Length = 215 Score = 256 bits (653), Expect = 9e-68 Identities = 128/186 (68%), Positives = 152/186 (81%) Frame = +1 Query: 16 NTHTMQNRQVQFKLVLLGESAVGKSSLVMRFVKGQFHEYQESTVGAAFLTQTVVVEDTTV 195 N ++ QFKLVLLGESAVGKSSLV+RFVKGQFHEYQEST+GAAFLTQ+V ++DTTV Sbjct: 10 NGQPQASKICQFKLVLLGESAVGKSSLVLRFVKGQFHEYQESTIGAAFLTQSVCLDDTTV 69 Query: 196 KFEIWDTAGQERYHSLAPMYYRGAQAAIVVYDITSQESFHRAKTWIQELTEKATSVKVIA 375 KFEIWDTAGQERYHSLAPMYYRGAQAAIVVYDIT+QE+F RAKTW++EL +A+ VIA Sbjct: 70 KFEIWDTAGQERYHSLAPMYYRGAQAAIVVYDITNQETFARAKTWVKELQRQASPSIVIA 129 Query: 376 LAGNKSDLSAQREVSISDGSTYAQENGLLFMETSAKTSSNVNEIFLEIAKKLPRHEVTTP 555 LAGNK+DL+ +R V + YA +N LLFMETSAKT+ NVN++FL IAKKLP+ E Sbjct: 130 LAGNKADLANKRMVEYEEAQAYADDNSLLFMETSAKTAMNVNDLFLAIAKKLPKSEPQNL 189 Query: 556 NAGSGQ 573 +G+ Sbjct: 190 GGAAGR 195
>sp|P29687|RAB5_TOBAC Ras-related protein Rab5 Length = 200 Score = 228 bits (580), Expect = 3e-59 Identities = 114/178 (64%), Positives = 139/178 (78%) Frame = +1 Query: 31 QNRQVQFKLVLLGESAVGKSSLVMRFVKGQFHEYQESTVGAAFLTQTVVVEDTTVKFEIW 210 ++ + KLVLLG+ GKSSLV+RFVKGQF E+QEST+GAAF + TV V + TVKFEIW Sbjct: 5 RHNNLNAKLVLLGDMGAGKSSLVIRFVKGQFLEFQESTIGAAFFSSTVSVNNATVKFEIW 64 Query: 211 DTAGQERYHSLAPMYYRGAQAAIVVYDITSQESFHRAKTWIQELTEKATSVKVIALAGNK 390 DTAGQERYHSLAPMYYRGA AAI+VYDITS ES RAK W+QEL ++ V+ALAGNK Sbjct: 65 DTAGQERYHSLAPMYYRGAAAAIIVYDITSTESLARAKKWVQELQKQGNPNMVMALAGNK 124 Query: 391 SDLSAQREVSISDGSTYAQENGLLFMETSAKTSSNVNEIFLEIAKKLPRHEVTTPNAG 564 +DL +R+V+ + YA+ENGL FMETSAKT++NVN+IF EIAK+LPR + AG Sbjct: 125 ADLEDKRKVTAEEARLYAEENGLFFMETSAKTATNVNDIFYEIAKRLPRAQPAQNPAG 182
>sp|P31582|RHA1_ARATH Ras-related protein RHA1 Length = 200 Score = 226 bits (577), Expect = 6e-59 Identities = 112/168 (66%), Positives = 133/168 (79%) Frame = +1 Query: 34 NRQVQFKLVLLGESAVGKSSLVMRFVKGQFHEYQESTVGAAFLTQTVVVEDTTVKFEIWD 213 N+ + KLVLLG+ GKSSLV+RFVK QF E+QEST+GAAF +QT+ V D TVKFEIWD Sbjct: 6 NKNINAKLVLLGDVGAGKSSLVLRFVKDQFVEFQESTIGAAFFSQTLAVNDATVKFEIWD 65 Query: 214 TAGQERYHSLAPMYYRGAQAAIVVYDITSQESFHRAKTWIQELTEKATSVKVIALAGNKS 393 TAGQERYHSLAPMYYRGA AAI+V+DIT+Q SF RAK W+QEL + V+ALAGNK+ Sbjct: 66 TAGQERYHSLAPMYYRGAAAAIIVFDITNQASFERAKKWVQELQAQGNPNMVMALAGNKA 125 Query: 394 DLSAQREVSISDGSTYAQENGLLFMETSAKTSSNVNEIFLEIAKKLPR 537 DL R+VS + YAQEN L FMETSAKT++NV +IF EIAK+LPR Sbjct: 126 DLLDARKVSAEEAEIYAQENSLFFMETSAKTATNVKDIFYEIAKRLPR 173
>sp|P31583|RHN1_NICPL Ras-related protein RHN1 Length = 200 Score = 226 bits (576), Expect = 8e-59 Identities = 113/171 (66%), Positives = 135/171 (78%) Frame = +1 Query: 52 KLVLLGESAVGKSSLVMRFVKGQFHEYQESTVGAAFLTQTVVVEDTTVKFEIWDTAGQER 231 KLVLLG+ GKSSLV+RFVKGQF E+QEST+GAAF + T+ V + TVKFEIWDTAGQER Sbjct: 12 KLVLLGDMGAGKSSLVIRFVKGQFLEFQESTIGAAFFSSTLAVNNATVKFEIWDTAGQER 71 Query: 232 YHSLAPMYYRGAQAAIVVYDITSQESFHRAKTWIQELTEKATSVKVIALAGNKSDLSAQR 411 YHSLAPMYYRGA AAI+VYDITS +SF RAK W+QEL ++ V+ALAGNK+DL +R Sbjct: 72 YHSLAPMYYRGAAAAIIVYDITSSDSFARAKKWVQELQKQGNPNMVMALAGNKADLEDRR 131 Query: 412 EVSISDGSTYAQENGLLFMETSAKTSSNVNEIFLEIAKKLPRHEVTTPNAG 564 +V+ + YA+ENGL FMETSAKT+ NVN IF EIAK+LPR + AG Sbjct: 132 KVTAEEARLYAEENGLFFMETSAKTAVNVNAIFYEIAKRLPRAQPAQNPAG 182
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 113,882,716
Number of Sequences: 369166
Number of extensions: 2312152
Number of successful extensions: 7486
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6769
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7085
length of database: 68,354,980
effective HSP length: 111
effective length of database: 47,849,395
effective search space used: 10670415085
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail