DrC_00748
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
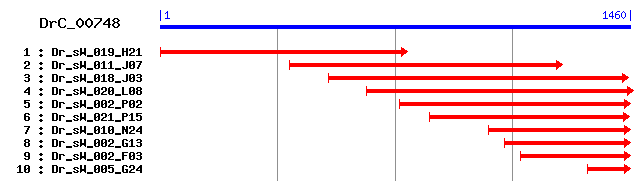
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00748 (1460 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q11067|PDIA6_CAEEL Probable protein disulfide-isomerase ... 491 e-138 sp|Q63081|PDIA6_RAT Protein disulfide-isomerase A6 precurso... 483 e-136 sp|Q922R8|PDIA6_MOUSE Protein disulfide-isomerase A6 precur... 482 e-135 sp|Q15084|PDIA6_HUMAN Protein disulfide-isomerase A6 precur... 480 e-135 sp|Q5R6T1|PDIA6_PONPY Protein disulfide-isomerase A6 precursor 477 e-134 sp|P38660|PDIA6_MESAU Protein disulfide-isomerase A6 precur... 471 e-132 sp|O22263|PDIA6_ARATH Probable protein disulfide-isomerase ... 177 9e-44 sp|P34329|PDIA4_CAEEL Probable protein disulfide-isomerase ... 163 1e-39 sp|Q92249|ERP38_NEUCR Protein disulfide-isomerase erp38 pre... 154 5e-37 sp|Q00216|TIGA_ASPNG Protein disulfide-isomerase tigA precu... 145 3e-34
>sp|Q11067|PDIA6_CAEEL Probable protein disulfide-isomerase A6 precursor Length = 440 Score = 491 bits (1265), Expect = e-138 Identities = 227/424 (53%), Positives = 304/424 (71%), Gaps = 2/424 (0%) Frame = +3 Query: 69 LYSPSDDVIDLTPSNFND-VKNGDELWFVEFYAPWCGHCQSLAPEWKKAASALKGVVKVG 245 +YS DDV++LT +NF V N D++W VEFYAPWCGHC+SL PE+KKAASALKGV KVG Sbjct: 19 MYSKKDDVVELTEANFQSKVINSDDIWIVEFYAPWCGHCKSLVPEYKKAASALKGVAKVG 78 Query: 246 AVDMDKYSSVGQPYNIQGFPTIKVFGKNKHSPKDYQGERNANAIVNNALKELEELVRSRL 425 AVDM ++ SVG PYN+QGFPT+K+FG +K P DY G+R A AI ++ L E ++ V +RL Sbjct: 79 AVDMTQHQSVGGPYNVQGFPTLKIFGADKKKPTDYNGQRTAQAIADSVLAEAKKAVSARL 138 Query: 426 XXXXXXXXXXXXXXXXXXXXXXXXX-EVIELTDSNFDELVLKSEEPWLVEFFAPWCGHCK 602 EV+ELTD+NF++LVL S++ WLVEFFAPWCGHCK Sbjct: 139 GGKSSGSSSSGSGSGSGKRGGGGSGNEVVELTDANFEDLVLNSKDIWLVEFFAPWCGHCK 198 Query: 603 NLKPHWDQAASELRGKVKLGALDATVHNAVASRYGVQGYPTIKFFPAGVKTGEAQEYDGG 782 +L+P W AASEL+GKV+LGALDATVH VA+++ ++G+PTIK+F G +AQ+YDGG Sbjct: 199 SLEPQWKAAASELKGKVRLGALDATVHTVVANKFAIRGFPTIKYFAPGSDVSDAQDYDGG 258 Query: 783 RSSSDIVRWASDKVVEQLPAPELKEITNQDVLKSTCEDHQLCVIAVLPNILDCQSACRNS 962 R SSDIV WAS + E +PAPE+ E NQ V++ C++ QLC+ A LP+ILDCQS CRN+ Sbjct: 259 RQSSDIVAWASARAQENMPAPEVFEGINQQVVEDACKEKQLCIFAFLPHILDCQSECRNN 318 Query: 963 YLDTIRVSSDKYKANRWGWCWTEAMKLPAIEKAFGIGGFGYPALVVVNVRKMKYATLTGP 1142 YL ++ S+K+K N WGW W E PA+E++F +GGFGYPA+ +N RK KYA L G Sbjct: 319 YLAMLKEQSEKFKKNLWGWIWVEGAAQPALEESFEVGGFGYPAMTALNFRKNKYAVLKGS 378 Query: 1143 FSQTGIQDFVRDVSLGRGSTAAIVGEGLPAITPVDPWDGKDGEMPKEEEIDLSELGIEEP 1322 F + GI +F+RD+S G+G T+++ G+G P I + WDGKDG +P E++IDLS++ ++ Sbjct: 379 FGKDGIHEFLRDLSYGKGRTSSLRGDGFPKIQKTEKWDGKDGALPAEDDIDLSDIDLD-- 436 Query: 1323 KSEL 1334 K+EL Sbjct: 437 KTEL 440
>sp|Q63081|PDIA6_RAT Protein disulfide-isomerase A6 precursor (Protein disulfide isomerase P5) (Calcium-binding protein 1) (CaBP1) (Thioredoxin domain-containing protein 7) Length = 440 Score = 483 bits (1244), Expect = e-136 Identities = 240/429 (55%), Positives = 299/429 (69%), Gaps = 6/429 (1%) Frame = +3 Query: 66 ALYSPSDDVIDLTPSNFN-DVKNGDELWFVEFYAPWCGHCQSLAPEWKKAASALKGVVKV 242 ALYS SDDVI+LTPSNFN +V D LW VEFYAPWCGHCQ L PEWKKAASALK VVKV Sbjct: 19 ALYSSSDDVIELTPSNFNREVIQSDSLWLVEFYAPWCGHCQRLTPEWKKAASALKDVVKV 78 Query: 243 GAVDMDKYSSVGQPYNIQGFPTIKVFGKNKHSPKDYQGERNANAIVNNALKELEELVRSR 422 GAV+ DK+ S+G Y +QGFPTIK+FG NK+ P+DYQG R AIV+ AL L +LV+ R Sbjct: 79 GAVNADKHQSLGGQYGVQGFPTIKIFGANKNKPEDYQGGRTGEAIVDAALSALRQLVKDR 138 Query: 423 LXXXXXXXXXXXXXXXXXXXXXXXXXEVIELTDSNFDELVLKSEEPWLVEFFAPWCGHCK 602 L +V+ELTD FD+ VL SE+ W+VEF+APWCGHCK Sbjct: 139 LGGRSGGYSSGKQGRGDSSSKK----DVVELTDDTFDKNVLDSEDVWMVEFYAPWCGHCK 194 Query: 603 NLKPHWDQAASELR----GKVKLGALDATVHNAVASRYGVQGYPTIKFFPAGVKTGEAQE 770 NL+P W AA+E++ GKVKL A+DATV+ +ASRYG++G+PTIK F G + Sbjct: 195 NLEPEWAAAATEVKEQTKGKVKLAAVDATVNQVLASRYGIKGFPTIKIFQKGESP---VD 251 Query: 771 YDGGRSSSDIVRWASDKVVEQLPAPELKEITNQDVLKSTCEDHQLCVIAVLPNILDCQSA 950 YDGGR+ SDIV A D + P PEL EI N+D+ K TCE+HQLCV+AVLP+ILD + Sbjct: 252 YDGGRTRSDIVSRALDLFSDNAPPPELLEIINEDIAKKTCEEHQLCVVAVLPHILDTGAT 311 Query: 951 CRNSYLDTIRVSSDKYKANRWGWCWTEAMKLPAIEKAFGIGGFGYPALVVVNVRKMKYAT 1130 RNSYL+ + +DKYK WGW WTEA +E A GIGGFGYPA+ +N RKMK+A Sbjct: 312 GRNSYLEVLLKLADKYKKKMWGWLWTEAGAQYELENALGIGGFGYPAMAAINARKMKFAL 371 Query: 1131 LTGPFSQTGIQDFVRDVSLGRGSTAAIVGEGLPAITPVDPWDGKDGEMPKEEEIDLSELG 1310 L G FS+ GI +F+R++S GRGSTA + G P ITP +PWDGKDGE+P E++IDLS++ Sbjct: 372 LKGSFSEQGINEFLRELSFGRGSTAPVGGGSFPNITprePWDGKDGELPVEDDIDLSDVE 431 Query: 1311 IEE-PKSEL 1334 +++ K EL Sbjct: 432 LDDLEKDEL 440
>sp|Q922R8|PDIA6_MOUSE Protein disulfide-isomerase A6 precursor (Thioredoxin domain-containing protein 7) Length = 440 Score = 482 bits (1241), Expect = e-135 Identities = 239/428 (55%), Positives = 299/428 (69%), Gaps = 6/428 (1%) Frame = +3 Query: 69 LYSPSDDVIDLTPSNFN-DVKNGDELWFVEFYAPWCGHCQSLAPEWKKAASALKGVVKVG 245 LYS SDDVI+LTPSNFN +V D LW VEFYAPWCGHCQ L PEWKKAA+ALK VVKVG Sbjct: 20 LYSSSDDVIELTPSNFNREVIQSDGLWLVEFYAPWCGHCQRLTPEWKKAATALKDVVKVG 79 Query: 246 AVDMDKYSSVGQPYNIQGFPTIKVFGKNKHSPKDYQGERNANAIVNNALKELEELVRSRL 425 AV+ DK+ S+G Y +QGFPTIK+FG NK+ P+DYQG R AIV+ AL L +LV+ RL Sbjct: 80 AVNADKHQSLGGQYGVQGFPTIKIFGANKNKPEDYQGGRTGEAIVDAALSALRQLVKDRL 139 Query: 426 XXXXXXXXXXXXXXXXXXXXXXXXXEVIELTDSNFDELVLKSEEPWLVEFFAPWCGHCKN 605 +V+ELTD FD+ VL SE+ W+VEF+APWCGHCKN Sbjct: 140 GGRSGGYSSGKQGRGDSSSKK----DVVELTDDTFDKNVLDSEDVWMVEFYAPWCGHCKN 195 Query: 606 LKPHWDQAASELR----GKVKLGALDATVHNAVASRYGVQGYPTIKFFPAGVKTGEAQEY 773 L+P W AA+E++ GKVKL A+DATV+ +ASRYG++G+PTIK F G +Y Sbjct: 196 LEPEWAAAATEVKEQTKGKVKLAAVDATVNQVLASRYGIKGFPTIKIFQKGESP---VDY 252 Query: 774 DGGRSSSDIVRWASDKVVEQLPAPELKEITNQDVLKSTCEDHQLCVIAVLPNILDCQSAC 953 DGGR+ SDIV A D + P PEL EI N+D+ K TCE+HQLCV+AVLP+ILD +A Sbjct: 253 DGGRTRSDIVSRALDLFSDNAPPPELLEIINEDIAKKTCEEHQLCVVAVLPHILDTGAAG 312 Query: 954 RNSYLDTIRVSSDKYKANRWGWCWTEAMKLPAIEKAFGIGGFGYPALVVVNVRKMKYATL 1133 RNSYL+ + +DKYK WGW WTEA +E A GIGGFGYPA+ +N RKMK+A L Sbjct: 313 RNSYLEVLLKLADKYKKKMWGWLWTEAGAQYELENALGIGGFGYPAMAAINARKMKFALL 372 Query: 1134 TGPFSQTGIQDFVRDVSLGRGSTAAIVGEGLPAITPVDPWDGKDGEMPKEEEIDLSELGI 1313 G FS+ GI +F+R++S GRGSTA + G P ITP +PWDGKDGE+P E++IDLS++ + Sbjct: 373 KGSFSEQGINEFLRELSFGRGSTAPVGGGSFPTITprePWDGKDGELPVEDDIDLSDVEL 432 Query: 1314 EE-PKSEL 1334 ++ K EL Sbjct: 433 DDLEKDEL 440
>sp|Q15084|PDIA6_HUMAN Protein disulfide-isomerase A6 precursor (Protein disulfide isomerase P5) (Thioredoxin domain-containing protein 7) Length = 440 Score = 480 bits (1235), Expect = e-135 Identities = 236/422 (55%), Positives = 295/422 (69%), Gaps = 5/422 (1%) Frame = +3 Query: 69 LYSPSDDVIDLTPSNFN-DVKNGDELWFVEFYAPWCGHCQSLAPEWKKAASALKGVVKVG 245 LYS SDDVI+LTPSNFN +V D LW VEFYAPWCGHCQ L PEWKKAA+ALK VVKVG Sbjct: 20 LYSSSDDVIELTPSNFNREVIQSDSLWLVEFYAPWCGHCQRLTPEWKKAATALKDVVKVG 79 Query: 246 AVDMDKYSSVGQPYNIQGFPTIKVFGKNKHSPKDYQGERNANAIVNNALKELEELVRSRL 425 AVD DK+ S+G Y +QGFPTIK+FG NK+ P+DYQG R AIV+ AL L +LV+ RL Sbjct: 80 AVDADKHHSLGGQYGVQGFPTIKIFGSNKNRPEDYQGGRTGEAIVDAALSALRQLVKDRL 139 Query: 426 XXXXXXXXXXXXXXXXXXXXXXXXXEVIELTDSNFDELVLKSEEPWLVEFFAPWCGHCKN 605 +VIELTD +FD+ VL SE+ W+VEF+APWCGHCKN Sbjct: 140 GGRSGGYSSGKQGRSDSSSKK----DVIELTDDSFDKNVLDSEDVWMVEFYAPWCGHCKN 195 Query: 606 LKPHWDQAASELR----GKVKLGALDATVHNAVASRYGVQGYPTIKFFPAGVKTGEAQEY 773 L+P W AASE++ GKVKL A+DATV+ +ASRYG++G+PTIK F G +Y Sbjct: 196 LEPEWAAAASEVKEQTKGKVKLAAVDATVNQVLASRYGIRGFPTIKIFQKGESP---VDY 252 Query: 774 DGGRSSSDIVRWASDKVVEQLPAPELKEITNQDVLKSTCEDHQLCVIAVLPNILDCQSAC 953 DGGR+ SDIV A D + P PEL EI N+D+ K TCE+HQLCV+AVLP+ILD +A Sbjct: 253 DGGRTRSDIVSRALDLFSDNAPPPELLEIINEDIAKRTCEEHQLCVVAVLPHILDTGAAG 312 Query: 954 RNSYLDTIRVSSDKYKANRWGWCWTEAMKLPAIEKAFGIGGFGYPALVVVNVRKMKYATL 1133 RNSYL+ + +DKYK WGW WTEA +E A GIGGFGYPA+ +N RKMK+A L Sbjct: 313 RNSYLEVLLKLADKYKKKMWGWLWTEAGAQSELETALGIGGFGYPAMAAINARKMKFALL 372 Query: 1134 TGPFSQTGIQDFVRDVSLGRGSTAAIVGEGLPAITPVDPWDGKDGEMPKEEEIDLSELGI 1313 G FS+ GI +F+R++S GRGSTA + G P I +PWDG+DGE+P E++IDLS++ + Sbjct: 373 KGSFSEQGINEFLRELSFGRGSTAPVGGGAFPTIVEREPWDGRDGELPVEDDIDLSDVEL 432 Query: 1314 EE 1319 ++ Sbjct: 433 DD 434
>sp|Q5R6T1|PDIA6_PONPY Protein disulfide-isomerase A6 precursor Length = 440 Score = 477 bits (1227), Expect = e-134 Identities = 234/422 (55%), Positives = 295/422 (69%), Gaps = 5/422 (1%) Frame = +3 Query: 69 LYSPSDDVIDLTPSNFN-DVKNGDELWFVEFYAPWCGHCQSLAPEWKKAASALKGVVKVG 245 LYS SDDVI+LTPSNFN +V D LW VEFYAPWCGHCQ L PEWKKAA+ALK VVKVG Sbjct: 20 LYSSSDDVIELTPSNFNREVIQSDSLWLVEFYAPWCGHCQRLTPEWKKAATALKDVVKVG 79 Query: 246 AVDMDKYSSVGQPYNIQGFPTIKVFGKNKHSPKDYQGERNANAIVNNALKELEELVRSRL 425 AVD DK+ S+G Y +QGFPTIK+FG NK+ P+DYQG R AIV+ AL L +LV+ RL Sbjct: 80 AVDADKHHSLGGQYGVQGFPTIKIFGSNKNRPEDYQGGRTGEAIVDAALSALRQLVKDRL 139 Query: 426 XXXXXXXXXXXXXXXXXXXXXXXXXEVIELTDSNFDELVLKSEEPWLVEFFAPWCGHCKN 605 +VIELTD +FD+ VL SE+ W+VEF+APWCGHCKN Sbjct: 140 GGQSGGYSSGKQGRSDSSSKK----DVIELTDDSFDKNVLDSEDVWMVEFYAPWCGHCKN 195 Query: 606 LKPHWDQAASELR----GKVKLGALDATVHNAVASRYGVQGYPTIKFFPAGVKTGEAQEY 773 L+P W AASE++ GKVKL A+DATV+ +ASRYG++G+PTIK F G +Y Sbjct: 196 LEPEWAAAASEVKEQTKGKVKLAAVDATVNQVLASRYGIRGFPTIKIFQKGESP---VDY 252 Query: 774 DGGRSSSDIVRWASDKVVEQLPAPELKEITNQDVLKSTCEDHQLCVIAVLPNILDCQSAC 953 DGGR+ SDIV A D + P PEL EI ++D+ K TCE+HQLCV++VLP+ILD +A Sbjct: 253 DGGRTRSDIVSRALDLFSDNAPPPELLEIISEDIAKRTCEEHQLCVVSVLPHILDTGAAG 312 Query: 954 RNSYLDTIRVSSDKYKANRWGWCWTEAMKLPAIEKAFGIGGFGYPALVVVNVRKMKYATL 1133 RNSYL+ + +DKYK WGW WTEA +E A GIGGFGYPA+ +N RKMK+A L Sbjct: 313 RNSYLEVLLKLADKYKKKMWGWLWTEAGAQSELETALGIGGFGYPAMAAINARKMKFALL 372 Query: 1134 TGPFSQTGIQDFVRDVSLGRGSTAAIVGEGLPAITPVDPWDGKDGEMPKEEEIDLSELGI 1313 G FS+ GI +F+R++S GRGSTA + G P I +PWDG+DGE+P E++IDLS++ + Sbjct: 373 KGSFSEQGINEFLRELSFGRGSTAPVGGGAFPTIVEREPWDGRDGELPVEDDIDLSDVEL 432 Query: 1314 EE 1319 ++ Sbjct: 433 DD 434
>sp|P38660|PDIA6_MESAU Protein disulfide-isomerase A6 precursor (Protein disulfide isomerase P5) Length = 439 Score = 471 bits (1212), Expect = e-132 Identities = 238/429 (55%), Positives = 299/429 (69%), Gaps = 7/429 (1%) Frame = +3 Query: 69 LYSPSDDVIDLTPSNFN-DVKNGDELWFVEFYAPWCGHCQSLAPEWKKAASALKGVVKVG 245 LYS SDDVI+LTPSNFN +V + LW VEFYAPWCGHCQ L PEWKKAA+ALK VVKVG Sbjct: 20 LYSSSDDVIELTPSNFNREVIQSNSLWLVEFYAPWCGHCQRLTPEWKKAATALKDVVKVG 79 Query: 246 AVDMDKYSSVGQPYNIQGFPTIKVFGKNKHSPKDYQGERNANAIVNNALKELEELVRSRL 425 AVD DK+ S+G Y +QGFPTIK+FG NK+ P+DYQG R AIV+ AL L +LV+ RL Sbjct: 80 AVDADKHQSLGGQYGVQGFPTIKIFGANKNKPEDYQGGRTGEAIVDAALSALRQLVKDRL 139 Query: 426 XXXXXXXXXXXXXXXXXXXXXXXXXEVIELTDSNFDELVLKSEEPWLVEFFAPWCGHCKN 605 +VIELTD FD+ VL S++ W+VEF+APWCGHCKN Sbjct: 140 SGRSGGYSSGKQGRGDSSSKK----DVIELTDDTFDKNVLDSDDVWMVEFYAPWCGHCKN 195 Query: 606 LKPHWDQAASELR----GKVKLGALDATVHNAVASRYGVQGYPTIKFFPAGVKTGEAQ-E 770 L+P W AA+E++ GKVKL A+DATV+ +A+RYG++G+PTIK F + GEA + Sbjct: 196 LEPEWATAATEVKEQTKGKVKLAAVDATVNQVLANRYGIRGFPTIKIF----QKGEAPVD 251 Query: 771 YDGGRSSSDIVRWASDKVVEQLPAPELKEITNQDVLKSTCEDHQLCVIAVLPNILDCQSA 950 YDGGR+ SDIV A D + P PEL EI N+DV K CE+HQLCV+AVLP+ILD A Sbjct: 252 YDGGRTRSDIVSRALDLFSDNAPPPELLEIINEDVAKKMCEEHQLCVVAVLPHILDT-GA 310 Query: 951 CRNSYLDTIRVSSDKYKANRWGWCWTEAMKLPAIEKAFGIGGFGYPALVVVNVRKMKYAT 1130 RNSYL+ + +DKYK WGW WTEA +E A GIGGFGYPA+ +N RKMK+A Sbjct: 311 ARNSYLEILLKLADKYKKKMWGWLWTEAGAQSELENALGIGGFGYPAMARINARKMKFAL 370 Query: 1131 LTGPFSQTGIQDFVRDVSLGRGSTAAIVGEGLPAITPVDPWDGKDGEMPKEEEIDLSELG 1310 L G FS+ GI +F+R++S GR STA + G PAIT +PWDG+DGE+P E++IDLS++ Sbjct: 371 LKGSFSEQGINEFLRELSFGRASTAPVGGGSFPAITAREPWDGRDGELPVEDDIDLSDVE 430 Query: 1311 IEE-PKSEL 1334 +++ K EL Sbjct: 431 LDDLEKDEL 439
>sp|O22263|PDIA6_ARATH Probable protein disulfide-isomerase A6 precursor (P5) Length = 361 Score = 177 bits (448), Expect = 9e-44 Identities = 97/251 (38%), Positives = 135/251 (53%), Gaps = 4/251 (1%) Frame = +3 Query: 81 SDDVIDLTPSNFNDVKNGDELWFVEFYAPWCGHCQSLAPEWKKAASALKGV--VKVGAVD 254 +DDV+ LT +F D+ VEFYAPWCGHC+ LAPE++K ++ K V + VD Sbjct: 22 ADDVVVLTDDSFEKEVGKDKGALVEFYAPWCGHCKKLAPEYEKLGASFKKAKSVLIAKVD 81 Query: 255 MDKYSSVGQPYNIQGFPTIKVFGKNKHSPKDYQGERNANAIVNNALKELEELVRSRLXXX 434 D+ SV Y + G+PTI+ F K P+ Y+G RNA A+ KE V+ Sbjct: 82 CDEQKSVCTKYGVSGYPTIQWFPKGSLEPQKYEGPRNAEALAEYVNKEGGTNVK------ 135 Query: 435 XXXXXXXXXXXXXXXXXXXXXXEVIELTDSNFDELVLKSEEPWLVEFFAPWCGHCKNLKP 614 V+ LT NFDE+VL + LVEF+APWCGHCK+L P Sbjct: 136 ----------------LAAVPQNVVVLTPDNFDEIVLDQNKDVLVEFYAPWCGHCKSLAP 179 Query: 615 HWDQAASELRGK--VKLGALDATVHNAVASRYGVQGYPTIKFFPAGVKTGEAQEYDGGRS 788 +++ A+ + + V + LDA H A+ +YGV G+PT+KFFP K G +YDGGR Sbjct: 180 TYEKVATVFKQEEGVVIANLDADAHKALGEKYGVSGFPTLKFFPKDNKAG--HDYDGGRD 237 Query: 789 SSDIVRWASDK 821 D V + ++K Sbjct: 238 LDDFVSFINEK 248
>sp|P34329|PDIA4_CAEEL Probable protein disulfide-isomerase A4 precursor (ERp-72 homolog) Length = 618 Score = 163 bits (413), Expect = 1e-39 Identities = 99/273 (36%), Positives = 149/273 (54%), Gaps = 7/273 (2%) Frame = +3 Query: 63 EALYSPSDDVIDLTPSNFNDVKNGDELWFVEFYAPWCGHCQSLAPEWKKAASALKGVVKV 242 E Y + V+ LT NF+ + V+FYAPWCGHC+ LAPE++KA+S K + + Sbjct: 29 ELNYEMDEGVVVLTDKNFDAFLKKNPSVLVKFYAPWCGHCKHLAPEYEKASS--KVSIPL 86 Query: 243 GAVDMDKYSSVGQPYNIQGFPTIKVFGKNKHSPKDYQGERNANAIVNNALKELEELVRSR 422 VD + +G+ + IQG+PT+K F K+ P DY G R+ IV E V SR Sbjct: 87 AKVDATVETELGKRFEIQGYPTLK-FWKDGKGPNDYDGGRDEAGIV--------EWVESR 137 Query: 423 LXXXXXXXXXXXXXXXXXXXXXXXXXEVIELTDSNFDELVLKSEEPWLVEFFAPWCGHCK 602 + EV+ LT NFD+ + + E LVEF+APWCGHCK Sbjct: 138 V----------------DPNYKPPPEEVVTLTTENFDDFI-SNNELVLVEFYAPWCGHCK 180 Query: 603 NLKPHWDQAASELR---GKVKLGALDATVHNAVASRYGVQGYPTIKFFPAGVKTGEAQEY 773 L P +++AA +L+ KVKLG +DAT+ + ++YGV GYPT+K ++ G +Y Sbjct: 181 KLAPEYEKAAQKLKAQGSKVKLGKVDATIEKDLGTKYGVSGYPTMKI----IRNGRRFDY 236 Query: 774 DGGRSSSDIVRWASDKVVEQLPA----PELKEI 860 +G R ++ I+++ +D + PA P+LK++ Sbjct: 237 NGpreAAGIIKYMTD---QSKPAAKKLPKLKDV 266
Score = 100 bits (248), Expect = 1e-20
Identities = 72/234 (30%), Positives = 108/234 (46%), Gaps = 7/234 (2%)
Frame = +3
Query: 504 VIELTDSNFDELVLKSEEPWLVEFFAPWCGHCKNLKPHWDQAASELRGKVKLGALDATVH 683
V+ LTD NFD LK LV+F+APWCGHCK+L P +++A+S++ + L +DATV
Sbjct: 38 VVVLTDKNFDAF-LKKNPSVLVKFYAPWCGHCKHLAPEYEKASSKV--SIPLAKVDATVE 94
Query: 684 NAVASRYGVQGYPTIKFFPAGVKTGEAQEYDGGRSSSDIVRWASDKVVEQL--PAPELKE 857
+ R+ +QGYPT+KF+ G + YDGGR + IV W +V P E+
Sbjct: 95 TELGKRFEIQGYPTLKFWKDGKGPND---YDGGRDEAGIVEWVESRVDPNYKPPPEEVVT 151
Query: 858 ITNQDVLKSTCEDHQLCVIAVLPNILDCQSAC--RNSYLDTIRVSSDKYKANRWGWCWTE 1031
+T ++ + + V P C+ ++ K K +
Sbjct: 152 LTTENFDDFISNNELVLVEFYAPWCGHCKKLAPEYEKAAQKLKAQGSKVKLGKVD----- 206
Query: 1032 AMKLPAIEKAFG--IGGFGYPALVVV-NVRKMKYATLTGPFSQTGIQDFVRDVS 1184
IEK G G GYP + ++ N R+ Y GP GI ++ D S
Sbjct: 207 ----ATIEKDLGTKYGVSGYPTMKIIRNGRRFDY---NGpreAAGIIKYMTDQS 253
Score = 77.8 bits (190), Expect = 7e-14
Identities = 40/99 (40%), Positives = 62/99 (62%), Gaps = 3/99 (3%)
Frame = +3
Query: 522 SNFDELVLKSEEPWLVEFFAPWCGHCKNLKPHW---DQAASELRGKVKLGALDATVHNAV 692
SNFD++V + L+EF+APWCGHCK+ + + QA + + V L +DAT+++A
Sbjct: 507 SNFDKIVNDESKDVLIEFYAPWCGHCKSFESKYVELAQALKKTQPNVVLAKMDATINDA- 565
Query: 693 ASRYGVQGYPTIKFFPAGVKTGEAQEYDGGRSSSDIVRW 809
S++ V+G+PTI F PAG K E +Y G R D+ ++
Sbjct: 566 PSQFAVEGFPTIYFAPAG-KKSEPIKYSGNRDLEDLKKF 603
Score = 54.7 bits (130), Expect = 7e-07
Identities = 37/104 (35%), Positives = 52/104 (50%), Gaps = 7/104 (6%)
Frame = +3
Query: 75 SPSDD---VIDLTPSNFNDVKNGDEL-WFVEFYAPWCGHCQSLAPEWKKAASALKGV-VK 239
+P DD V + SNF+ + N + +EFYAPWCGHC+S ++ + A ALK
Sbjct: 493 APKDDKGPVKTVVGSNFDKIVNDESKDVLIEFYAPWCGHCKSFESKYVELAQALKKTQPN 552
Query: 240 VGAVDMD-KYSSVGQPYNIQGFPTIKVFGKNKHS-PKDYQGERN 365
V MD + + ++GFPTI K S P Y G R+
Sbjct: 553 VVLAKMDATINDAPSQFAVEGFPTIYFAPAGKKSEPIKYSGNRD 596
>sp|Q92249|ERP38_NEUCR Protein disulfide-isomerase erp38 precursor (ERp38) Length = 369 Score = 154 bits (390), Expect = 5e-37 Identities = 88/253 (34%), Positives = 137/253 (54%), Gaps = 9/253 (3%) Frame = +3 Query: 90 VIDLTPSNFNDV--KNGDELWFVEFYAPWCGHCQSLAPEWKKAASAL---KGVVKVGAVD 254 V+DL PSNF+DV K+G VEF+APWCGHC++LAP +++ A+AL K V++ VD Sbjct: 22 VLDLIPSNFDDVVLKSGKPT-LVEFFAPWCGHCKNLAPVYEELATALEYAKDKVQIAKVD 80 Query: 255 MDKYSSVGQPYNIQGFPTIKVFGKNKHSPKDYQGERNANAIVNNALKELEELVRSRLXXX 434 D ++G+ + +QGFPT+K F P DY+G R+ +++ N ++ R + Sbjct: 81 ADAERALGKRFGVQGFPTLKFFDGKSEQPVDYKGGRDLDSLSNFIAEKTGVKARKK---- 136 Query: 435 XXXXXXXXXXXXXXXXXXXXXXEVIELTDSNFDELVLKSEEPWLVEFFAPWCGHCKNLKP 614 ++ + + + + ++ LV F APWCGHCKNL P Sbjct: 137 ------------------GSAPSLVNILNDATIKGAIGGDKNVLVAFTAPWCGHCKNLAP 178 Query: 615 HWDQAASELRGKVKLGA----LDATVHNAVASRYGVQGYPTIKFFPAGVKTGEAQEYDGG 782 W++ A+ ++ DA A+ YGV G+PTIKFFP G T E +Y+GG Sbjct: 179 TWEKLAATFASDPEITIAKVDADAPTGKKSAAEYGVSGFPTIKFFPKGSTTPE--DYNGG 236 Query: 783 RSSSDIVRWASDK 821 RS +D+V++ ++K Sbjct: 237 RSEADLVKFLNEK 249
Score = 107 bits (268), Expect = 7e-23
Identities = 56/133 (42%), Positives = 83/133 (62%), Gaps = 7/133 (5%)
Frame = +3
Query: 504 VIELTDSNFDELVLKSEEPWLVEFFAPWCGHCKNLKPHWDQAASEL---RGKVKLGALDA 674
V++L SNFD++VLKS +P LVEFFAPWCGHCKNL P +++ A+ L + KV++ +DA
Sbjct: 22 VLDLIPSNFDDVVLKSGKPTLVEFFAPWCGHCKNLAPVYEELATALEYAKDKVQIAKVDA 81
Query: 675 TVHNAVASRYGVQGYPTIKFFPAGVKTGEAQEYDGGRSSSDIVRWASD----KVVEQLPA 842
A+ R+GVQG+PT+KFF K+ + +Y GGR + + ++ K ++ A
Sbjct: 82 DAERALGKRFGVQGFPTLKFFDG--KSEQPVDYKGGRDLDSLSNFIAEKTGVKARKKGSA 139
Query: 843 PELKEITNQDVLK 881
P L I N +K
Sbjct: 140 PSLVNILNDATIK 152
>sp|Q00216|TIGA_ASPNG Protein disulfide-isomerase tigA precursor Length = 359 Score = 145 bits (366), Expect = 3e-34 Identities = 91/253 (35%), Positives = 128/253 (50%), Gaps = 9/253 (3%) Frame = +3 Query: 90 VIDLTPSNFNDV--KNGDELWFVEFYAPWCGHCQSLAPEWKKAASALKGV---VKVGAVD 254 V+DL P NF+DV K+G VEF+APWCGHC++LAP +++ A V VG VD Sbjct: 21 VVDLVPKNFDDVVLKSGKPA-LVEFFAPWCGHCKNLAPVYEELGQAFAHASDKVTVGKVD 79 Query: 255 MDKYSSVGQPYNIQGFPTIKVFGKNKHSPKDYQGERNANAIVNNALKELEELVRSRLXXX 434 D++ +G+ + +QGFPT+K F P+DY+G R+ L+ L + + Sbjct: 80 ADEHRDLGRKFGVQGFPTLKWFDGKSDEPEDYKGGRD--------LESLSSFISEK---- 127 Query: 435 XXXXXXXXXXXXXXXXXXXXXXEVIELTDSNFDELVLKSEEPWLVEFFAPWCGHCKNLKP 614 +V L D+ F V + LV F APWCGHCKNL P Sbjct: 128 ---------TGVKPRGPKKEPSKVEMLNDATFKGAV-GGDNDVLVAFTAPWCGHCKNLAP 177 Query: 615 HWDQAASE--LRGKVKLGALDATVHN--AVASRYGVQGYPTIKFFPAGVKTGEAQEYDGG 782 W+ A++ L V + +DA N A A GV GYPTIKFFP G + E+ Y+G Sbjct: 178 TWEALANDFVLEPNVVIAKVDADAENGKATAREQGVSGYPTIKFFPKG--STESVPYEGA 235 Query: 783 RSSSDIVRWASDK 821 RS + + ++K Sbjct: 236 RSEQAFIDFLNEK 248
Score = 104 bits (259), Expect = 7e-22
Identities = 80/255 (31%), Positives = 119/255 (46%), Gaps = 14/255 (5%)
Frame = +3
Query: 504 VIELTDSNFDELVLKSEEPWLVEFFAPWCGHCKNLKPHWD---QAASELRGKVKLGALDA 674
V++L NFD++VLKS +P LVEFFAPWCGHCKNL P ++ QA + KV +G +DA
Sbjct: 21 VVDLVPKNFDDVVLKSGKPALVEFFAPWCGHCKNLAPVYEELGQAFAHASDKVTVGKVDA 80
Query: 675 TVHNAVASRYGVQGYPTIKFFPAGVKTGEAQEYDGGRSSSDIVRWASDKVVEQLPAPELK 854
H + ++GVQG+PT+K+F K+ E ++Y GGR + + S+K + P+ +
Sbjct: 81 DEHRDLGRKFGVQGFPTLKWFDG--KSDEPEDYKGGRDLESLSSFISEKTGVKPRGPKKE 138
Query: 855 ----EITNQDVLKSTC-EDHQLCVIAVLP------NILDCQSACRNSYLDTIRVSSDKYK 1001
E+ N K D+ + V P N+ A N ++ V K
Sbjct: 139 PSKVEMLNDATFKGAVGGDNDVLVAFTAPWCGHCKNLAPTWEALANDFVLEPNVVIAKVD 198
Query: 1002 ANRWGWCWTEAMKLPAIEKAFGIGGFGYPALVVVNVRKMKYATLTGPFSQTGIQDFVRDV 1181
A+ E K A E+ G GYP + + G S+ DF+ +
Sbjct: 199 AD------AENGKATAREQ----GVSGYPTIKFFPKGSTESVPYEGARSEQAFIDFLNE- 247
Query: 1182 SLGRGSTAAIVGEGL 1226
+ T VG GL
Sbjct: 248 ---KTGTHRTVGGGL 259
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 159,779,831
Number of Sequences: 369166
Number of extensions: 3238647
Number of successful extensions: 10495
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9211
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10183
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 17477978805
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail