DrC_00368
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
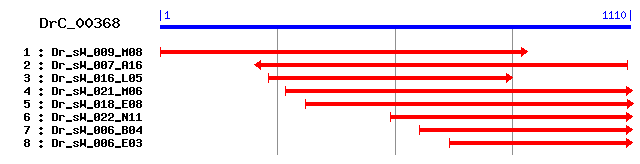
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00368 (1110 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P52209|6PGD_HUMAN 6-phosphogluconate dehydrogenase, deca... 473 e-133 sp|P00349|6PGD_SHEEP 6-phosphogluconate dehydrogenase, deca... 473 e-133 sp|Q9DCD0|6PGD_MOUSE 6-phosphogluconate dehydrogenase, deca... 469 e-132 sp|P41570|6PGD_CERCA 6-phosphogluconate dehydrogenase, deca... 453 e-127 sp|P41572|6PGD_DROME 6-phosphogluconate dehydrogenase, deca... 444 e-124 sp|Q17761|6PGD_CAEEL 6-phosphogluconate dehydrogenase, deca... 441 e-123 sp|P41573|6PGD_DROSI 6-phosphogluconate dehydrogenase, deca... 441 e-123 sp|O60037|6PGD_CUNEL 6-phosphogluconate dehydrogenase, deca... 441 e-123 sp|P78812|6PGD_SCHPO 6-phosphogluconate dehydrogenase, deca... 426 e-119 sp|P38720|6PGD1_YEAST 6-phosphogluconate dehydrogenase, dec... 417 e-116
>sp|P52209|6PGD_HUMAN 6-phosphogluconate dehydrogenase, decarboxylating Length = 483 Score = 473 bits (1217), Expect = e-133 Identities = 225/353 (63%), Positives = 271/353 (76%) Frame = +3 Query: 3 GEEGARFGPSLMPGGNVEAWKSIQPLFQDIAAKANKVEPCCDWVGSDGAGHFVKMVHNGI 182 GEEGAR+GPSLMPGGN EAW I+ +FQ IAAK EPCCDWVG +GAGHFVKMVHNGI Sbjct: 131 GEEGARYGPSLMPGGNKEAWPHIKTIFQGIAAKVGTGEPCCDWVGDEGAGHFVKMVHNGI 190 Query: 183 EYGDMQLICEAYHLMRDLLEMGNEEISNAFADWNNSELESFLIEITSSITKFKANDGTYL 362 EYGDMQLICEAYHLM+D+L M +E++ AF DWN +EL+SFLIEIT++I KF+ DG +L Sbjct: 191 EYGDMQLICEAYHLMKDVLGMAQDEMAQAFEDWNKTELDSFLIEITANILKFQDTDGKHL 250 Query: 363 VEDIYDSAGQKGTGKWTAITALDFGVPVTLIAESVFARSLSALKSLRVEASKRFPSNNLL 542 + I DSAGQKGTGKWTAI+AL++GVPVTLI E+VFAR LS+LK R++ASK+ Sbjct: 251 LPKIRDSAGQKGTGKWTAISALEYGVPVTLIGEAVFARCLSSLKDERIQASKKLKGPQKF 310 Query: 543 NFTGDKTKFLNQIKYALYASKIISYTQGFMMMKSASHEFKWNLDLGKIALMWRGGCIIRS 722 F GDK FL I+ ALYASKIISY QGFM+++ A+ EF W L+ G IALMWRGGCIIRS Sbjct: 311 QFDGDKKSFLEDIRKALYASKIISYAQGFMLLRQAATEFGWTLNYGGIALMWRGGCIIRS 370 Query: 723 RFLQNIKDAYEKNKNLSILLFDKFFQSEILKCQESWRHVVANAALHGIPTPAFSCALAFY 902 FL IKDA+++N L LL D FF+S + CQ+SWR V+ GIP P F+ AL+FY Sbjct: 371 VFLGKIKDAFDRNPELQNLLLDDFFKSAVENCQDSWRRAVSTGVQAGIPMPCFTTALSFY 430 Query: 903 DGLRTEKLPANLIQAQRDLFGAHQYQLLDHPGTYEHTDWTGHGGNAAASTYQA 1061 DG R E LPA+LIQAQRD FGAH Y+LL PG + HT+WTGHGG ++S+Y A Sbjct: 431 DGYRHEMLPASLIQAQRDYFGAHTYELLAKPGQFIHTNWTGHGGTVSSSSYNA 483
>sp|P00349|6PGD_SHEEP 6-phosphogluconate dehydrogenase, decarboxylating Length = 483 Score = 473 bits (1216), Expect = e-133 Identities = 224/353 (63%), Positives = 274/353 (77%) Frame = +3 Query: 3 GEEGARFGPSLMPGGNVEAWKSIQPLFQDIAAKANKVEPCCDWVGSDGAGHFVKMVHNGI 182 GE+GAR+GPSLMPGGN EAW I+ +FQ IAAK EPCCDWVG DGAGHFVKMVHNGI Sbjct: 131 GEDGARYGPSLMPGGNKEAWPHIKAIFQGIAAKVGTGEPCCDWVGDDGAGHFVKMVHNGI 190 Query: 183 EYGDMQLICEAYHLMRDLLEMGNEEISNAFADWNNSELESFLIEITSSITKFKANDGTYL 362 EYGDMQLICEAYHLM+D+L +G++E++ AF +WN +EL+SFLIEIT+SI KF+ DG +L Sbjct: 191 EYGDMQLICEAYHLMKDVLGLGHKEMAKAFEEWNKTELDSFLIEITASILKFQDADGKHL 250 Query: 363 VEDIYDSAGQKGTGKWTAITALDFGVPVTLIAESVFARSLSALKSLRVEASKRFPSNNLL 542 + I DSAGQKGTGKWTAI+AL++GVPVTLI E+VFAR LS+LK R++ASK+ + Sbjct: 251 LPKIRDSAGQKGTGKWTAISALEYGVPVTLIGEAVFARCLSSLKDERIQASKKLKGPQNI 310 Query: 543 NFTGDKTKFLNQIKYALYASKIISYTQGFMMMKSASHEFKWNLDLGKIALMWRGGCIIRS 722 F GDK FL I+ ALYASKIISY QGFM+++ A+ EF W L+ G IALMWRGGCIIRS Sbjct: 311 PFEGDKKSFLEDIRKALYASKIISYAQGFMLLRQAATEFGWTLNYGGIALMWRGGCIIRS 370 Query: 723 RFLQNIKDAYEKNKNLSILLFDKFFQSEILKCQESWRHVVANAALHGIPTPAFSCALAFY 902 FL IKDA+++N L LL D FF+S + CQ+SWR ++ GIP P F+ AL+FY Sbjct: 371 VFLGKIKDAFDRNPGLQNLLLDDFFKSAVENCQDSWRRAISTGVQAGIPMPCFTTALSFY 430 Query: 903 DGLRTEKLPANLIQAQRDLFGAHQYQLLDHPGTYEHTDWTGHGGNAAASTYQA 1061 DG R LPANLIQAQRD FGAH Y+LL PG + HT+WTGHGG+ ++S+Y A Sbjct: 431 DGYRHAMLPANLIQAQRDYFGAHTYELLAKPGQFIHTNWTGHGGSVSSSSYNA 483
>sp|Q9DCD0|6PGD_MOUSE 6-phosphogluconate dehydrogenase, decarboxylating Length = 483 Score = 469 bits (1206), Expect = e-132 Identities = 223/353 (63%), Positives = 273/353 (77%) Frame = +3 Query: 3 GEEGARFGPSLMPGGNVEAWKSIQPLFQDIAAKANKVEPCCDWVGSDGAGHFVKMVHNGI 182 GEEGAR+GPSLMPGGN EAW I+ +FQ IAAK EPCCDWVG +GAGHFVKMVHNGI Sbjct: 131 GEEGARYGPSLMPGGNKEAWPHIKAIFQAIAAKVGTGEPCCDWVGDEGAGHFVKMVHNGI 190 Query: 183 EYGDMQLICEAYHLMRDLLEMGNEEISNAFADWNNSELESFLIEITSSITKFKANDGTYL 362 EYGDMQLICEAYHLM+D+L M +EE++ AF +WN +EL+SFLIEIT++I K++ DG L Sbjct: 191 EYGDMQLICEAYHLMKDVLGMRHEEMAQAFEEWNKTELDSFLIEITANILKYRDTDGKEL 250 Query: 363 VEDIYDSAGQKGTGKWTAITALDFGVPVTLIAESVFARSLSALKSLRVEASKRFPSNNLL 542 + I DSAGQKGTGKWTAI+AL++G+PVTLI E+VFAR LS+LK RV+AS++ ++ Sbjct: 251 LPKIRDSAGQKGTGKWTAISALEYGMPVTLIGEAVFARCLSSLKEERVQASQKLKGPKVV 310 Query: 543 NFTGDKTKFLNQIKYALYASKIISYTQGFMMMKSASHEFKWNLDLGKIALMWRGGCIIRS 722 G K FL I+ ALYASKIISY QGFM+++ A+ EF W L+ G IALMWRGGCIIRS Sbjct: 311 QLEGSKKSFLEDIRKALYASKIISYAQGFMLLRQAATEFGWTLNYGGIALMWRGGCIIRS 370 Query: 723 RFLQNIKDAYEKNKNLSILLFDKFFQSEILKCQESWRHVVANAALHGIPTPAFSCALAFY 902 FL IKDA+E+N L LL D FF+S + CQ+SWR V++ GIP P F+ AL+FY Sbjct: 371 VFLGKIKDAFERNPELQNLLLDDFFKSAVDNCQDSWRRVISTGVQAGIPMPCFTTALSFY 430 Query: 903 DGLRTEKLPANLIQAQRDLFGAHQYQLLDHPGTYEHTDWTGHGGNAAASTYQA 1061 DG R E LPANLIQAQRD FGAH Y+LL PG + HT+WTGHGG+ ++S+Y A Sbjct: 431 DGYRHEMLPANLIQAQRDYFGAHTYELLTKPGEFIHTNWTGHGGSVSSSSYNA 483
>sp|P41570|6PGD_CERCA 6-phosphogluconate dehydrogenase, decarboxylating Length = 481 Score = 453 bits (1165), Expect = e-127 Identities = 231/353 (65%), Positives = 267/353 (75%) Frame = +3 Query: 3 GEEGARFGPSLMPGGNVEAWKSIQPLFQDIAAKANKVEPCCDWVGSDGAGHFVKMVHNGI 182 GEEGAR GPSLMPGG+ EAW IQP+FQ I AKA+K EPCC+WVG GAGHFVKMVHNGI Sbjct: 132 GEEGARHGPSLMPGGHPEAWPLIQPIFQSICAKADK-EPCCEWVGEGGAGHFVKMVHNGI 190 Query: 183 EYGDMQLICEAYHLMRDLLEMGNEEISNAFADWNNSELESFLIEITSSITKFKANDGTYL 362 EYGDMQLICEAY +M+ L + E++ F WN+ EL+SFLIEIT I ++ + G YL Sbjct: 191 EYGDMQLICEAYQIMK-ALGLSQAEMATEFEKWNSEELDSFLIEITRDILNYQDDRG-YL 248 Query: 363 VEDIYDSAGQKGTGKWTAITALDFGVPVTLIAESVFARSLSALKSLRVEASKRFPSNNLL 542 +E I D+AGQKGTGKWTAI+AL +GVPVTLI E+VF+R LSALK RV ASK+ N+ Sbjct: 249 LERIRDTAGQKGTGKWTAISALQYGVPVTLIGEAVFSRCLSALKDERVAASKQLKGPNVN 308 Query: 543 NFTGDKTKFLNQIKYALYASKIISYTQGFMMMKSASHEFKWNLDLGKIALMWRGGCIIRS 722 D KFLN IK+ALY SKI+SY QGFM+M+ A+ E WNL+ G IALMWRGGCIIRS Sbjct: 309 AKVEDLPKFLNHIKHALYCSKIVSYAQGFMLMREAAKENNWNLNYGGIALMWRGGCIIRS 368 Query: 723 RFLQNIKDAYEKNKNLSILLFDKFFQSEILKCQESWRHVVANAALHGIPTPAFSCALAFY 902 FL NIKDAY +N LS LL D FF+ I Q SWR VVANA L GIP PA S AL+FY Sbjct: 369 VFLGNIKDAYTRNPQLSNLLLDDFFKKAIEVGQNSWRQVVANAFLWGIPVPALSTALSFY 428 Query: 903 DGLRTEKLPANLIQAQRDLFGAHQYQLLDHPGTYEHTDWTGHGGNAAASTYQA 1061 DG RTEKLPANL+QAQRD FGAH Y+LL G + HT+WTG GGN +ASTYQA Sbjct: 429 DGYRTEKLPANLLQAQRDYFGAHTYELLGAEGKFVHTNWTGTGGNVSASTYQA 481
>sp|P41572|6PGD_DROME 6-phosphogluconate dehydrogenase, decarboxylating Length = 481 Score = 444 bits (1141), Expect = e-124 Identities = 224/353 (63%), Positives = 265/353 (75%) Frame = +3 Query: 3 GEEGARFGPSLMPGGNVEAWKSIQPLFQDIAAKANKVEPCCDWVGSDGAGHFVKMVHNGI 182 GEEGAR GPSLMPGG+ AW IQP+FQ I AKA+ EPCC+WVG GAGHFVKMVHNGI Sbjct: 132 GEEGARHGPSLMPGGHEAAWPLIQPIFQAICAKADG-EPCCEWVGDGGAGHFVKMVHNGI 190 Query: 183 EYGDMQLICEAYHLMRDLLEMGNEEISNAFADWNNSELESFLIEITSSITKFKANDGTYL 362 EYGDMQLICEAYH+M+ L + +++++ F WN++EL+SFLIEIT I K+K G YL Sbjct: 191 EYGDMQLICEAYHIMKSL-GLSADQMADEFGKWNSAELDSFLIEITRDILKYKDGKG-YL 248 Query: 363 VEDIYDSAGQKGTGKWTAITALDFGVPVTLIAESVFARSLSALKSLRVEASKRFPSNNLL 542 +E I D+AGQKGTGKWTAI AL +GVPVTLI E+VF+R LSALK RV+AS + Sbjct: 249 LERIRDTAGQKGTGKWTAIAALQYGVPVTLIGEAVFSRCLSALKDERVQASSVLKGPSTK 308 Query: 543 NFTGDKTKFLNQIKYALYASKIISYTQGFMMMKSASHEFKWNLDLGKIALMWRGGCIIRS 722 + TKFL+ IK+ALY +KI+SY QGFM+M+ A+ E KW L+ G IALMWRGGCIIRS Sbjct: 309 AQVANLTKFLDDIKHALYCAKIVSYAQGFMLMREAARENKWRLNYGGIALMWRGGCIIRS 368 Query: 723 RFLQNIKDAYEKNKNLSILLFDKFFQSEILKCQESWRHVVANAALHGIPTPAFSCALAFY 902 FL NIKDAY LS LL D FF+ I + Q+SWR VVANA GIP PA S AL+FY Sbjct: 369 VFLGNIKDAYTSQPELSNLLLDDFFKKAIERGQDSWREVVANAFRWGIPVPALSTALSFY 428 Query: 903 DGLRTEKLPANLIQAQRDLFGAHQYQLLDHPGTYEHTDWTGHGGNAAASTYQA 1061 DG RT KLPANL+QAQRD FGAH Y+LL G + HT+WTG GGN +ASTYQA Sbjct: 429 DGYRTAKLPANLLQAQRDYFGAHTYELLGQEGQFHHTNWTGTGGNVSASTYQA 481
>sp|Q17761|6PGD_CAEEL 6-phosphogluconate dehydrogenase, decarboxylating Length = 484 Score = 441 bits (1135), Expect = e-123 Identities = 218/355 (61%), Positives = 264/355 (74%), Gaps = 2/355 (0%) Frame = +3 Query: 3 GEEGARFGPSLMPGGNVEAWKSIQPLFQDIAAKANKVEPCCDWVGSDGAGHFVKMVHNGI 182 GEEGARFGPSLMPGGN +AW ++ +FQ IAAK+N EPCCDWVG+ G+GHFVKMVHNGI Sbjct: 131 GEEGARFGPSLMPGGNPKAWPHLKDIFQKIAAKSNG-EPCCDWVGNAGSGHFVKMVHNGI 189 Query: 183 EYGDMQLICEAYHLMRDLLEMGNEEISNAFADWNNSELESFLIEITSSITKFKANDGTYL 362 EYGDMQLI EAYHL+ +E+ +++++ DWN ELESFLIEIT++I K++ G + Sbjct: 190 EYGDMQLIAEAYHLLSKAVELNHDQMAEVLDDWNKGELESFLIEITANILKYRDEQGEPI 249 Query: 363 VEDIYDSAGQKGTGKWTAITALDFGVPVTLIAESVFARSLSALKSLRVEASKRFPSNNLL 542 V I DSAGQKGTGKWT AL++G+PVTLI E+VFAR LSALK RV ASK+ P + Sbjct: 250 VPKIRDSAGQKGTGKWTCFAALEYGLPVTLIGEAVFARCLSALKDERVRASKQLPRPQVS 309 Query: 543 NFT--GDKTKFLNQIKYALYASKIISYTQGFMMMKSASHEFKWNLDLGKIALMWRGGCII 716 T DK F+ QI ALYASKI+SY QGFM++ AS +F WNL+ G IALMWRGGCII Sbjct: 310 PDTVVQDKRVFIKQISKALYASKIVSYAQGFMLLAEASKQFNWNLNFGAIALMWRGGCII 369 Query: 717 RSRFLQNIKDAYEKNKNLSILLFDKFFQSEILKCQESWRHVVANAALHGIPTPAFSCALA 896 RSRFL +I+ A++KNK LS LL D FF I + Q+SWR VV A GIP PAFS ALA Sbjct: 370 RSRFLGDIEHAFQKNKQLSNLLLDDFFTKAITEAQDSWRVVVCAAVRLGIPVPAFSSALA 429 Query: 897 FYDGLRTEKLPANLIQAQRDLFGAHQYQLLDHPGTYEHTDWTGHGGNAAASTYQA 1061 FYDG +E +PANL+QAQRD FGAH Y+LL PGT+ HT+WTG GG ++ Y A Sbjct: 430 FYDGYTSEVVPANLLQAQRDYFGAHTYELLAKPGTWVHTNWTGTGGRVTSNAYNA 484
>sp|P41573|6PGD_DROSI 6-phosphogluconate dehydrogenase, decarboxylating Length = 481 Score = 441 bits (1135), Expect = e-123 Identities = 223/353 (63%), Positives = 265/353 (75%) Frame = +3 Query: 3 GEEGARFGPSLMPGGNVEAWKSIQPLFQDIAAKANKVEPCCDWVGSDGAGHFVKMVHNGI 182 GEEGAR GPSLMPGG+ AW IQP+FQ I AKA+ EPCC+WVG GAGHFVKMVHNGI Sbjct: 132 GEEGARHGPSLMPGGHEAAWPLIQPIFQAICAKADG-EPCCEWVGDGGAGHFVKMVHNGI 190 Query: 183 EYGDMQLICEAYHLMRDLLEMGNEEISNAFADWNNSELESFLIEITSSITKFKANDGTYL 362 EYGDMQLICEAYH+M+ L + +++++ F WN++EL+SFLIEIT I K+K G +L Sbjct: 191 EYGDMQLICEAYHIMQSL-GLSADQMADEFGKWNSAELDSFLIEITRDILKYKDGKG-HL 248 Query: 363 VEDIYDSAGQKGTGKWTAITALDFGVPVTLIAESVFARSLSALKSLRVEASKRFPSNNLL 542 +E I D+AGQKGTGKWTAI AL +GVPVTLI E+VF+R LSALK RV+AS + Sbjct: 249 LERIRDTAGQKGTGKWTAIAALQYGVPVTLIGEAVFSRCLSALKDERVQASSVLKGPSTK 308 Query: 543 NFTGDKTKFLNQIKYALYASKIISYTQGFMMMKSASHEFKWNLDLGKIALMWRGGCIIRS 722 + TKFL+ IK+ALY +KI+SY QGFM+M+ A+ E KW L+ G IALMWRGGCIIRS Sbjct: 309 AEVANLTKFLDDIKHALYCAKIVSYAQGFMLMREAARENKWRLNYGGIALMWRGGCIIRS 368 Query: 723 RFLQNIKDAYEKNKNLSILLFDKFFQSEILKCQESWRHVVANAALHGIPTPAFSCALAFY 902 FL NIKDAY LS LL D FF+ I + Q+SWR VVANA GIP PA S AL+FY Sbjct: 369 VFLGNIKDAYTSQPQLSNLLLDDFFKKAIERGQDSWREVVANAFRWGIPVPALSTALSFY 428 Query: 903 DGLRTEKLPANLIQAQRDLFGAHQYQLLDHPGTYEHTDWTGHGGNAAASTYQA 1061 DG RT KLPANL+QAQRD FGAH Y+LL G + HT+WTG GGN +ASTYQA Sbjct: 429 DGYRTAKLPANLLQAQRDYFGAHTYELLGQEGQFHHTNWTGTGGNVSASTYQA 481
>sp|O60037|6PGD_CUNEL 6-phosphogluconate dehydrogenase, decarboxylating Length = 485 Score = 441 bits (1134), Expect = e-123 Identities = 214/353 (60%), Positives = 263/353 (74%) Frame = +3 Query: 3 GEEGARFGPSLMPGGNVEAWKSIQPLFQDIAAKANKVEPCCDWVGSDGAGHFVKMVHNGI 182 GEEGAR+GPSLMPGGN +AW+ IQP+FQ IAAKA CC+WVG GAGH+VKMVHNGI Sbjct: 133 GEEGARYGPSLMPGGNSKAWEHIQPIFQAIAAKAPDGASCCEWVGETGAGHYVKMVHNGI 192 Query: 183 EYGDMQLICEAYHLMRDLLEMGNEEISNAFADWNNSELESFLIEITSSITKFKANDGTYL 362 EYGDMQLI E Y ++ + L + ++E+++ F +WN +L+SFLIEIT I +FK DG L Sbjct: 193 EYGDMQLITEVYQILHEGLGLSHDEMADIFEEWNKGDLDSFLIEITRDILRFKDTDGQPL 252 Query: 363 VEDIYDSAGQKGTGKWTAITALDFGVPVTLIAESVFARSLSALKSLRVEASKRFPSNNLL 542 V I D+AGQKGTGKWTAI +LD G+PVTLI E+V++R LS+LK RV ASK + Sbjct: 253 VTKIRDTAGQKGTGKWTAIDSLDRGIPVTLIGEAVYSRCLSSLKDERVRASKILQGPSSS 312 Query: 543 NFTGDKTKFLNQIKYALYASKIISYTQGFMMMKSASHEFKWNLDLGKIALMWRGGCIIRS 722 FTGDK F+ Q+ ALYA+KI+SY QG+M+M+ A+ +++W L+ IALMWRGGCIIRS Sbjct: 313 KFTGDKKTFIAQLGQALYAAKIVSYAQGYMLMRQAAKDYEWKLNNAGIALMWRGGCIIRS 372 Query: 723 RFLQNIKDAYEKNKNLSILLFDKFFQSEILKCQESWRHVVANAALHGIPTPAFSCALAFY 902 FL I+DAY KN L LLFD FF+ K Q++WR+V A A L GIPTPA S AL FY Sbjct: 373 VFLGKIRDAYTKNPELENLLFDDFFKDATAKAQDAWRNVTAQAVLMGIPTPALSTALNFY 432 Query: 903 DGLRTEKLPANLIQAQRDLFGAHQYQLLDHPGTYEHTDWTGHGGNAAASTYQA 1061 DGLR E LPANL+QAQRD FGAH Y+LL PG + HT+WTG GGN +ASTY A Sbjct: 433 DGLRHEILPANLLQAQRDYFGAHTYELLHTPGKWVHTNWTGRGGNVSASTYDA 485
>sp|P78812|6PGD_SCHPO 6-phosphogluconate dehydrogenase, decarboxylating Length = 492 Score = 426 bits (1094), Expect = e-119 Identities = 210/360 (58%), Positives = 261/360 (72%), Gaps = 7/360 (1%) Frame = +3 Query: 3 GEEGARFGPSLMPGGNVEAWKSIQPLFQDIAAKANKVEPCCDWVGSDGAGHFVKMVHNGI 182 GEEGAR+GPSLMPGGN AW I+P+FQ +AAKA EPCCDWVG GAGH+VKMVHNGI Sbjct: 134 GEEGARYGPSLMPGGNPAAWPRIKPIFQTLAAKAGNNEPCCDWVGEQGAGHYVKMVHNGI 193 Query: 183 EYGDMQLICEAYHLMRDLLEMGNEEISNAFADWNNSELESFLIEITSSITKFKANDGTYL 362 EYGDMQLICE Y +M+ L M +EI++ F WN +L+SFLIEIT + ++KA+DG L Sbjct: 194 EYGDMQLICETYDIMKRGLGMSCDEIADVFEKWNTGKLDSFLIEITRDVLRYKADDGKPL 253 Query: 363 VEDIYDSAGQKGTGKWTAITALDFGVPVTLIAESVFARSLSALKSLRVEASKRFPSNNLL 542 VE I D+AGQKGTGKWTA AL+ G PV+LI E+VFAR LS+LKS RV ASK+ N Sbjct: 254 VEKILDAAGQKGTGKWTAQNALEMGTPVSLITEAVFARCLSSLKSERVRASKKLTGPN-T 312 Query: 543 NFTGDKTKFLNQIKYALYASKIISYTQGFMMMKSASHEFKWNLDLGKIALMWRGGCIIRS 722 FTGDK + ++ ++ ALYASKIISY QGFM+M+ A+ E+ W L+ IALMWRGGCIIRS Sbjct: 313 KFTGDKKQLIDDLEDALYASKIISYAQGFMLMREAAKEYGWKLNNAGIALMWRGGCIIRS 372 Query: 723 RFLQNIKDAYEKNKNLSILLFDKFFQSEILKCQESWRHVVANAALHGIPTPAFSCALAFY 902 FL++I +A+ ++ NL +LF FF + + K Q WR VVA AA+ GIP PA S L+FY Sbjct: 373 VFLKDITEAFREDPNLESILFHPFFTNGVEKAQAGWRRVVAQAAMLGIPVPATSTGLSFY 432 Query: 903 DGLRTEKLPANLIQAQRDLFGAHQYQLLDHPGTYE-------HTDWTGHGGNAAASTYQA 1061 DG R+ LPANL+QAQRD FGAH +++L H +WTGHGGN +A+TY A Sbjct: 433 DGYRSAVLPANLLQAQRDYFGAHTFRVLPEAADKSLPADKDIHINWTGHGGNISATTYDA 492
>sp|P38720|6PGD1_YEAST 6-phosphogluconate dehydrogenase, decarboxylating 1 Length = 489 Score = 417 bits (1073), Expect = e-116 Identities = 211/361 (58%), Positives = 263/361 (72%), Gaps = 8/361 (2%) Frame = +3 Query: 3 GEEGARFGPSLMPGGNVEAWKSIQPLFQDIAAKANKVEPCCDWVGSDGAGHFVKMVHNGI 182 GEEGAR+GPSLMPGG+ EAW I+ +FQ I+AK++ EPCC+WVG GAGH+VKMVHNGI Sbjct: 130 GEEGARYGPSLMPGGSEEAWPHIKNIFQSISAKSDG-EPCCEWVGPAGAGHYVKMVHNGI 188 Query: 183 EYGDMQLICEAYHLMRDLLEMGNEEISNAFADWNNSELESFLIEITSSITKFKANDGTYL 362 EYGDMQLICEAY +M+ L ++EIS+ FA WNN L+SFL+EIT I KF DG L Sbjct: 189 EYGDMQLICEAYDIMKRLGGFTDKEISDVFAKWNNGVLDSFLVEITRDILKFDDVDGKPL 248 Query: 363 VEDIYDSAGQKGTGKWTAITALDFGVPVTLIAESVFARSLSALKSLRVEASKRFPSNNL- 539 VE I D+AGQKGTGKWTAI ALD G+PVTLI E+VFAR LSALK+ R+ ASK P + Sbjct: 249 VEKIMDTAGQKGTGKWTAINALDLGMPVTLIGEAVFARCLSALKNERIRASKVLPGPEVP 308 Query: 540 LNFTGDKTKFLNQIKYALYASKIISYTQGFMMMKSASHEFKWNLDLGKIALMWRGGCIIR 719 + D+ +F++ ++ ALYASKIISY QGFM+++ A+ + W L+ IALMWRGGCIIR Sbjct: 309 KDAVKDREQFVDDLEQALYASKIISYAQGFMLIREAAATYGWKLNNPAIALMWRGGCIIR 368 Query: 720 SRFLQNIKDAYEKNKNLSILLFDKFFQSEILKCQESWRHVVANAALHGIPTPAFSCALAF 899 S FL I AY + +L LLF+KFF + K Q WR +A A +GIPTPAFS AL+F Sbjct: 369 SVFLGQITKAYREEPDLENLLFNKFFADAVTKAQSGWRKSIALATTYGIPTPAFSTALSF 428 Query: 900 YDGLRTEKLPANLIQAQRDLFGAHQYQLLDHPGTYE-------HTDWTGHGGNAAASTYQ 1058 YDG R+E+LPANL+QAQRD FGAH +++L + H +WTGHGGN ++STYQ Sbjct: 429 YDGYRSERLPANLLQAQRDYFGAHTFRVLPECASDNLPVDKDIHINWTGHGGNVSSSTYQ 488 Query: 1059 A 1061 A Sbjct: 489 A 489
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 136,080,081
Number of Sequences: 369166
Number of extensions: 2991348
Number of successful extensions: 7867
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7404
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7765
length of database: 68,354,980
effective HSP length: 112
effective length of database: 47,664,660
effective search space used: 12249817620
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail