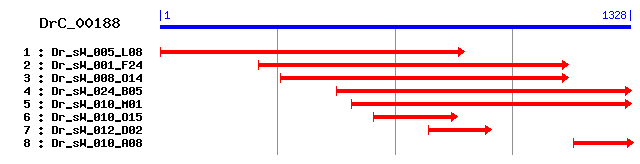
DrC_00188
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00188 (1328 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q8R5C5|ACTY_MOUSE Beta-centractin (Actin-related protein... 605 e-172 sp|P42025|ACTY_HUMAN Beta-centractin (Actin-related protein... 603 e-172 sp|P61164|ACTZ_MOUSE Alpha-centractin (Centractin) (Centros... 597 e-170 sp|P45889|ACTZ_DROME Actin-like protein 87C 581 e-165 sp|P42023|ACTZ_PNECA Actin-2 (Actin II) (Centractin-like pr... 489 e-138 sp|P38673|ACTZ_NEUCR Actin-like protein (Centractin) 462 e-129 sp|P02578|ACT1_ACACA Actin-1 426 e-118 sp|P53496|ACT11_ARATH Actin-11 426 e-118 sp|P10365|ACT_THELA Actin 425 e-118 sp|P53498|ACT_CHLRE Actin 424 e-118
>sp|Q8R5C5|ACTY_MOUSE Beta-centractin (Actin-related protein 1B) (ARP1B) Length = 376 Score = 605 bits (1559), Expect = e-172 Identities = 290/366 (79%), Positives = 327/366 (89%) Frame = +2 Query: 2 NQPVVIDNGSGVIKAGFAGATTPKIHFPNYIGRPKHKRVMAGALEGDTFIGPKAEEHRGL 181 NQPVVIDNGSGVIKAGFAG PK FPNY+GRPKH RVMAGALEGD FIGPKAEEHRGL Sbjct: 9 NQPVVIDNGSGVIKAGFAGDQIPKYCFPNYVGRPKHMRVMAGALEGDLFIGPKAEEHRGL 68 Query: 182 MNIRYPMEHGVVQDWNDMEKIWSYIYGPEQLKTSSEEHPVLLTEAPLNPLKNREKAAEIF 361 + IRYPMEHGVV+DWNDME+IW Y+Y +QL+T SEEHPVLLTEAPLNP KNREKAAE+F Sbjct: 69 LTIRYPMEHGVVRDWNDMERIWQYVYSKDQLQTFSEEHPVLLTEAPLNPSKNREKAAEVF 128 Query: 362 FETFNVPALYISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFALQHAIMRTDIAGR 541 FETFNVPAL+ISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFA+ H+IMR DIAGR Sbjct: 129 FETFNVPALFISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFAMPHSIMRVDIAGR 188 Query: 542 DVSRYLRLLLRKEGTHFQTSAEFEIVRQLKEKACFLSANPEMQEQSEIDNTTYPLPDGSK 721 DVSRYLRLLLRKEG F TSAEFE+VR +KE+AC+LS NP+ E E + Y LPDGS Sbjct: 189 DVSRYLRLLLRKEGADFHTSAEFEVVRTIKERACYLSINPQKDEALETEKVQYTLPDGST 248 Query: 722 LRIGGAKFRAPELLFRPDLIGSECDGIHEVLAYSIQKSDMDLRQILYGNIVLSGGSTLFK 901 L +G A+FRAPELLF+PDL+G E +G+HEVLA++I KSDMDLR+ L+ NIVLSGGSTLFK Sbjct: 249 LDVGPARFRAPELLFQPDLVGDESEGLHEVLAFAIHKSDMDLRRTLFSNIVLSGGSTLFK 308 Query: 902 GFGDRILSELKKLAPKDNKIRISAPQERLYSTWIGGSILASLDTFRRMWISKREFEDDGY 1081 GFGDR+LSE+KKLAPKD KI+ISAPQERLYSTWIGGSILASLDTF++MW+SK+E+E+DG Sbjct: 309 GFGDRLLSEVKKLAPKDVKIKISAPQERLYSTWIGGSILASLDTFKKMWVSKKEYEEDGS 368 Query: 1082 RALHRK 1099 RA+HRK Sbjct: 369 RAIHRK 374
>sp|P42025|ACTY_HUMAN Beta-centractin (Actin-related protein 1B) (ARP1B) Length = 376 Score = 603 bits (1556), Expect = e-172 Identities = 289/366 (78%), Positives = 327/366 (89%) Frame = +2 Query: 2 NQPVVIDNGSGVIKAGFAGATTPKIHFPNYIGRPKHKRVMAGALEGDTFIGPKAEEHRGL 181 NQPVVIDNGSGVIKAGFAG PK FPNY+GRPKH RVMAGALEGD FIGPKAEEHRGL Sbjct: 9 NQPVVIDNGSGVIKAGFAGDQIPKYCFPNYVGRPKHMRVMAGALEGDLFIGPKAEEHRGL 68 Query: 182 MNIRYPMEHGVVQDWNDMEKIWSYIYGPEQLKTSSEEHPVLLTEAPLNPLKNREKAAEIF 361 + IRYPMEHGVV+DWNDME+IW Y+Y +QL+T SEEHPVLLTEAPLNP KNREKAAE+F Sbjct: 69 LTIRYPMEHGVVRDWNDMERIWQYVYSKDQLQTFSEEHPVLLTEAPLNPSKNREKAAEVF 128 Query: 362 FETFNVPALYISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFALQHAIMRTDIAGR 541 FETFNVPAL+ISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFA+ H+IMR DIAGR Sbjct: 129 FETFNVPALFISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFAMPHSIMRVDIAGR 188 Query: 542 DVSRYLRLLLRKEGTHFQTSAEFEIVRQLKEKACFLSANPEMQEQSEIDNTTYPLPDGSK 721 DVSRYLRLLLRKEG F TSAEFE+VR +KE+AC+LS NP+ E E + Y LPDGS Sbjct: 189 DVSRYLRLLLRKEGVDFHTSAEFEVVRTIKERACYLSINPQKDEALETEKVQYTLPDGST 248 Query: 722 LRIGGAKFRAPELLFRPDLIGSECDGIHEVLAYSIQKSDMDLRQILYGNIVLSGGSTLFK 901 L +G A+FRAPELLF+PDL+G E +G+HEV+A++I KSDMDLR+ L+ NIVLSGGSTLFK Sbjct: 249 LDVGPARFRAPELLFQPDLVGDESEGLHEVVAFAIHKSDMDLRRTLFANIVLSGGSTLFK 308 Query: 902 GFGDRILSELKKLAPKDNKIRISAPQERLYSTWIGGSILASLDTFRRMWISKREFEDDGY 1081 GFGDR+LSE+KKLAPKD KI+ISAPQERLYSTWIGGSILASLDTF++MW+SK+E+E+DG Sbjct: 309 GFGDRLLSEVKKLAPKDIKIKISAPQERLYSTWIGGSILASLDTFKKMWVSKKEYEEDGS 368 Query: 1082 RALHRK 1099 RA+HRK Sbjct: 369 RAIHRK 374
>sp|P61164|ACTZ_MOUSE Alpha-centractin (Centractin) (Centrosome-associated actin homolog) (Actin-RPV) (ARP1) sp|P61163|ACTZ_HUMAN Alpha-centractin (Centractin) (Centrosome-associated actin homolog) (Actin-RPV) (ARP1) sp|P61162|ACTZ_CANFA Alpha-centractin (Centractin) (Centrosome-associated actin homolog) (Actin-RPV) (ARP1) Length = 376 Score = 597 bits (1538), Expect = e-170 Identities = 286/366 (78%), Positives = 327/366 (89%) Frame = +2 Query: 2 NQPVVIDNGSGVIKAGFAGATTPKIHFPNYIGRPKHKRVMAGALEGDTFIGPKAEEHRGL 181 NQPVVIDNGSGVIKAGFAG PK FPNY+GRPKH RVMAGALEGD FIGPKAEEHRGL Sbjct: 9 NQPVVIDNGSGVIKAGFAGDQIPKYCFPNYVGRPKHVRVMAGALEGDIFIGPKAEEHRGL 68 Query: 182 MNIRYPMEHGVVQDWNDMEKIWSYIYGPEQLKTSSEEHPVLLTEAPLNPLKNREKAAEIF 361 ++IRYPMEHG+V+DWNDME+IW Y+Y +QL+T SEEHPVLLTEAPLNP KNRE+AAE+F Sbjct: 69 LSIRYPMEHGIVKDWNDMERIWQYVYSKDQLQTFSEEHPVLLTEAPLNPRKNRERAAEVF 128 Query: 362 FETFNVPALYISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFALQHAIMRTDIAGR 541 FETFNVPAL+ISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFA+ H+IMR DIAGR Sbjct: 129 FETFNVPALFISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFAMPHSIMRIDIAGR 188 Query: 542 DVSRYLRLLLRKEGTHFQTSAEFEIVRQLKEKACFLSANPEMQEQSEIDNTTYPLPDGSK 721 DVSR+LRL LRKEG F +S+EFEIV+ +KE+AC+LS NP+ E E + Y LPDGS Sbjct: 189 DVSRFLRLYLRKEGYDFHSSSEFEIVKAIKERACYLSINPQKDETLETEKAQYYLPDGST 248 Query: 722 LRIGGAKFRAPELLFRPDLIGSECDGIHEVLAYSIQKSDMDLRQILYGNIVLSGGSTLFK 901 + IG ++FRAPELLFRPDLIG E +GIHEVL ++IQKSDMDLR+ L+ NIVLSGGSTLFK Sbjct: 249 IEIGPSRFRAPELLFRPDLIGEESEGIHEVLVFAIQKSDMDLRRTLFSNIVLSGGSTLFK 308 Query: 902 GFGDRILSELKKLAPKDNKIRISAPQERLYSTWIGGSILASLDTFRRMWISKREFEDDGY 1081 GFGDR+LSE+KKLAPKD KIRISAPQERLYSTWIGGSILASLDTF++MW+SK+E+E+DG Sbjct: 309 GFGDRLLSEVKKLAPKDVKIRISAPQERLYSTWIGGSILASLDTFKKMWVSKKEYEEDGA 368 Query: 1082 RALHRK 1099 R++HRK Sbjct: 369 RSIHRK 374
>sp|P45889|ACTZ_DROME Actin-like protein 87C Length = 376 Score = 581 bits (1497), Expect = e-165 Identities = 276/366 (75%), Positives = 321/366 (87%) Frame = +2 Query: 2 NQPVVIDNGSGVIKAGFAGATTPKIHFPNYIGRPKHKRVMAGALEGDTFIGPKAEEHRGL 181 NQPVVIDNGSGVIKAGFAG PK FPNYIGRPKH RVMAGALEGD F+GPKAEEHRGL Sbjct: 9 NQPVVIDNGSGVIKAGFAGEHIPKCRFPNYIGRPKHVRVMAGALEGDIFVGPKAEEHRGL 68 Query: 182 MNIRYPMEHGVVQDWNDMEKIWSYIYGPEQLKTSSEEHPVLLTEAPLNPLKNREKAAEIF 361 ++IRYPMEHG+V DWNDME+IWSYIY EQL T +E+HPVLLTEAPLNP +NREKAAE F Sbjct: 69 LSIRYPMEHGIVTDWNDMERIWSYIYSKEQLATFTEDHPVLLTEAPLNPRRNREKAAEFF 128 Query: 362 FETFNVPALYISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFALQHAIMRTDIAGR 541 FE N PAL++SMQAVLSLYATGR TGVVLDSGDGVTHAVPIYEGFA+ H+IMR DIAGR Sbjct: 129 FEGINAPALFVSMQAVLSLYATGRVTGVVLDSGDGVTHAVPIYEGFAMPHSIMRVDIAGR 188 Query: 542 DVSRYLRLLLRKEGTHFQTSAEFEIVRQLKEKACFLSANPEMQEQSEIDNTTYPLPDGSK 721 DV+RYL+ L+R+EG +F+++AEFEIVR +KEK C+L+ NP+ +E E + Y LPDG Sbjct: 189 DVTRYLKTLIRREGFNFRSTAEFEIVRSIKEKVCYLATNPQKEETVETEKFAYKLPDGKI 248 Query: 722 LRIGGAKFRAPELLFRPDLIGSECDGIHEVLAYSIQKSDMDLRQILYGNIVLSGGSTLFK 901 IG A+FRAPE+LFRPDL+G EC+GIH+VL YSI+KSDMDLR++LY NIVLSGGSTLFK Sbjct: 249 FEIGPARFRAPEVLFRPDLLGEECEGIHDVLMYSIEKSDMDLRKMLYQNIVLSGGSTLFK 308 Query: 902 GFGDRILSELKKLAPKDNKIRISAPQERLYSTWIGGSILASLDTFRRMWISKREFEDDGY 1081 GFGDR+LSELKK + KD KIRI+APQERLYSTW+GGSILASLDTF++MWISKRE+E++G Sbjct: 309 GFGDRLLSELKKHSAKDLKIRIAAPQERLYSTWMGGSILASLDTFKKMWISKREYEEEGQ 368 Query: 1082 RALHRK 1099 +A+HRK Sbjct: 369 KAVHRK 374
>sp|P42023|ACTZ_PNECA Actin-2 (Actin II) (Centractin-like protein) Length = 385 Score = 489 bits (1260), Expect = e-138 Identities = 243/377 (64%), Positives = 302/377 (80%), Gaps = 11/377 (2%) Frame = +2 Query: 2 NQPVVIDNGSGVIKAGFAGATTPKIHFPNYIGRPKHKRVMAGALEGDTFIGPKAEEHRGL 181 NQP+ IDNGSGVIKAGFAG PK FP+Y+GRPKH ++MAGA+EGD FIG KA+E RGL Sbjct: 9 NQPICIDNGSGVIKAGFAGEDQPKSFFPSYVGRPKHLKIMAGAIEGDIFIGNKAQELRGL 68 Query: 182 MNIRYPMEHGVVQDWNDMEKIWSYIYGPEQLKTSSEEHPVLLTEAPLNPLKNREKAAEIF 361 + I+YP+EHG+V DW+DME+IW +IY E+LKT SEEHPVLLTEAPLNP NR++AA++F Sbjct: 69 LKIKYPIEHGIVVDWDDMERIWQFIY-TEELKTVSEEHPVLLTEAPLNPRTNRDQAAQVF 127 Query: 362 FETFNVPALYISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFALQHAIMRTDIAGR 541 FETFNVPAL+ S+QAVLSLYA+GRTTGVVLDSGDGVTHAVPIYEGFA+ AI R DIAGR Sbjct: 128 FETFNVPALFTSIQAVLSLYASGRTTGVVLDSGDGVTHAVPIYEGFAMPSAIRRIDIAGR 187 Query: 542 DVSRYLRLLLRKEGTHFQTSAEFEIVRQLKEKACFLSANPEMQEQSEID----------- 688 DV+ YL+LLLRK GT F TSAE EIVR +KEK +++ +P +E+ I+ Sbjct: 188 DVTEYLQLLLRKSGTIFHTSAEKEIVRIIKEKCSYVTLDPRKEEKEWINASISGGKDYTK 247 Query: 689 NTTYPLPDGSKLRIGGAKFRAPELLFRPDLIGSECDGIHEVLAYSIQKSDMDLRQILYGN 868 + LPDG+ LR+G +FRAPE+LF P++IGSE GIH+V+ +I + D+DLR+ L+GN Sbjct: 248 EEEFKLPDGNVLRLGAERFRAPEILFDPEIIGSEYSGIHQVVVDAISRVDLDLRKSLFGN 307 Query: 869 IVLSGGSTLFKGFGDRILSELKKLAPKDNKIRISAPQERLYSTWIGGSILASLDTFRRMW 1048 IVLSGGSTL +GFGDR+LSE+++LA KD KI+I AP ER YSTWIGGSILASL TFR+MW Sbjct: 308 IVLSGGSTLTRGFGDRLLSEIRRLAVKDVKIKIFAPPERKYSTWIGGSILASLSTFRKMW 367 Query: 1049 ISKREFEDDGYRALHRK 1099 +S E+++D +HRK Sbjct: 368 VSAEEYQEDP-DIIHRK 383
>sp|P38673|ACTZ_NEUCR Actin-like protein (Centractin) Length = 380 Score = 462 bits (1188), Expect = e-129 Identities = 227/366 (62%), Positives = 291/366 (79%), Gaps = 8/366 (2%) Frame = +2 Query: 2 NQPVVIDNGSGVIKAGFAGATTPKIHFPNYIGRPKHKRVMAGALEGDTFIGPKA-EEHRG 178 N P+V+DNGSG I+AGFAG PK HFP+++GRPKH RV+AGALEG+ FIG KA E RG Sbjct: 7 NAPIVLDNGSGTIRAGFAGDDVPKCHFPSFVGRPKHLRVLAGALEGEVFIGQKAASELRG 66 Query: 179 LMNIRYPMEHGVVQDWNDMEKIWSYIYGPEQLKTSSEEHPVLLTEAPLNPLKNREKAAEI 358 L+ IRYP+EHG+V DW+DMEKIW+Y+Y E LKT SEEHPVLLTE PLNP NR+ AA+I Sbjct: 67 LLKIRYPLEHGIVTDWDDMEKIWAYVYD-EGLKTLSEEHPVLLTEPPLNPRANRDTAAQI 125 Query: 359 FFETFNVPALYISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFALQHAIMRTDIAG 538 FETFNVPALY S+QAVLSLYA+GRTTGVVLDSGDGV+HAVP+Y+GF + ++I R D+AG Sbjct: 126 LFETFNVPALYTSIQAVLSLYASGRTTGVVLDSGDGVSHAVPVYQGFTVPNSIRRIDVAG 185 Query: 539 RDVSRYLRLLLRKEGTHFQTSAEFEIVRQLKEKACFLSANPEMQEQ----SEIDN---TT 697 RDV+ YL+ LLRK G F TSAE E+VR +KE +++ +P +E+ +++D Sbjct: 186 RDVTEYLQTLLRKSGYVFHTSAEKEVVRLIKESVTYVAHDPRKEEKEWAAAKMDPAKIAE 245 Query: 698 YPLPDGSKLRIGGAKFRAPELLFRPDLIGSECDGIHEVLAYSIQKSDMDLRQILYGNIVL 877 Y LPDG+KL+IG +FRAPE+LF P++IG E G+H+++ SI ++D+DLR+ LY NIVL Sbjct: 246 YVLPDGNKLKIGAERFRAPEILFDPEIIGLEYPGVHQIVVDSINRTDLDLRRDLYSNIVL 305 Query: 878 SGGSTLFKGFGDRILSELKKLAPKDNKIRISAPQERLYSTWIGGSILASLDTFRRMWISK 1057 SGGSTL KGFGDR+L+E++KLA KD +I+I AP ER YSTWIGGSILA L TFR+MW+S Sbjct: 306 SGGSTLTKGFGDRLLTEVQKLAVKDMRIKIFAPPERKYSTWIGGSILAGLSTFRKMWVSI 365 Query: 1058 REFEDD 1075 ++ ++ Sbjct: 366 DDWHEN 371
>sp|P02578|ACT1_ACACA Actin-1 Length = 375 Score = 426 bits (1094), Expect = e-118 Identities = 205/369 (55%), Positives = 272/369 (73%), Gaps = 4/369 (1%) Frame = +2 Query: 5 QPVVIDNGSGVIKAGFAGATTPKIHFPNYIGRPKHKRVMAGALEGDTFIGPKAEEHRGLM 184 Q +VIDNGSG+ KAGFAG P+ FP+ +GRP+H VM G + D+++G +A+ RG++ Sbjct: 6 QALVIDNGSGMCKAGFAGDDAPRAVFPSIVGRPRHTGVMVGMGQKDSYVGDEAQSKRGIL 65 Query: 185 NIRYPMEHGVVQDWNDMEKIWSYIYGPEQLKTSSEEHPVLLTEAPLNPLKNREKAAEIFF 364 ++YP+EHG+V +W+DMEKIW + + E L+ + EEHPVLLTEAPLNP NREK +I F Sbjct: 66 TLKYPIEHGIVTNWDDMEKIWHHTFYNE-LRVAPEEHPVLLTEAPLNPKANREKMTQIMF 124 Query: 365 ETFNVPALYISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFALQHAIMRTDIAGRD 544 ETFN PA+Y+++QAVLSLYA+GRTTG+VLDSGDGVTH VPIYEG+AL HAI+R D+AGRD Sbjct: 125 ETFNTPAMYVAIQAVLSLYASGRTTGIVLDSGDGVTHTVPIYEGYALPHAILRLDLAGRD 184 Query: 545 VSRYLRLLLRKEGTHFQTSAEFEIVRQLKEKACFLSANPEMQEQSEIDNT----TYPLPD 712 ++ YL +L + G F T+AE EIVR +KEK C+++ + E + + ++ +Y LPD Sbjct: 185 LTDYLMKILTERGYSFTTTAEREIVRDIKEKLCYVALDFEQEMHTAASSSALEKSYELPD 244 Query: 713 GSKLRIGGAKFRAPELLFRPDLIGSECDGIHEVLAYSIQKSDMDLRQILYGNIVLSGGST 892 G + IG +FRAPE LF+P +G E GIHE SI K D+D+R+ LYGN+VLSGG+T Sbjct: 245 GQVITIGNERFRAPEALFQPSFLGMESAGIHETTYNSIMKCDVDIRKDLYGNVVLSGGTT 304 Query: 893 LFKGFGDRILSELKKLAPKDNKIRISAPQERLYSTWIGGSILASLDTFRRMWISKREFED 1072 +F G DR+ EL LAP KI+I AP ER YS WIGGSILASL TF++MWISK E+++ Sbjct: 305 MFPGIADRMQKELTALAPSTMKIKIIAPPERKYSVWIGGSILASLSTFQQMWISKEEYDE 364 Query: 1073 DGYRALHRK 1099 G +HRK Sbjct: 365 SGPSIVHRK 373
>sp|P53496|ACT11_ARATH Actin-11 Length = 377 Score = 426 bits (1094), Expect = e-118 Identities = 204/371 (54%), Positives = 274/371 (73%), Gaps = 6/371 (1%) Frame = +2 Query: 5 QPVVIDNGSGVIKAGFAGATTPKIHFPNYIGRPKHKRVMAGALEGDTFIGPKAEEHRGLM 184 QP+V DNG+G++KAGFAG P+ FP+ +GRP+H VM G + D ++G +A+ RG++ Sbjct: 8 QPLVCDNGTGMVKAGFAGDDAPRAVFPSIVGRPRHTGVMVGMGQKDAYVGDEAQSKRGIL 67 Query: 185 NIRYPMEHGVVQDWNDMEKIWSYIYGPEQLKTSSEEHPVLLTEAPLNPLKNREKAAEIFF 364 ++YP+EHG+V +W+DMEKIW + + E L+ + EEHPVLLTEAPLNP NREK +I F Sbjct: 68 TLKYPIEHGIVSNWDDMEKIWHHTFYNE-LRVAPEEHPVLLTEAPLNPKANREKMTQIMF 126 Query: 365 ETFNVPALYISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFALQHAIMRTDIAGRD 544 ETFN PA+Y+++QAVLSLYA+GRTTG+VLDSGDGV+H VPIYEG+AL HAI+R D+AGRD Sbjct: 127 ETFNTPAMYVAIQAVLSLYASGRTTGIVLDSGDGVSHTVPIYEGYALPHAILRLDLAGRD 186 Query: 545 VSRYLRLLLRKEGTHFQTSAEFEIVRQLKEKACFLSANPEMQEQSEIDNT------TYPL 706 ++ YL +L + G F TSAE EIVR +KEK +++ + +++ E NT +Y L Sbjct: 187 LTDYLMKILTERGYSFTTSAEREIVRDVKEKLAYIAL--DYEQEMETANTSSSVEKSYEL 244 Query: 707 PDGSKLRIGGAKFRAPELLFRPDLIGSECDGIHEVLAYSIQKSDMDLRQILYGNIVLSGG 886 PDG + IGG +FR PE+LF+P L+G E GIHE SI K D+D+R+ LYGNIVLSGG Sbjct: 245 PDGQVITIGGERFRCPEVLFQPSLVGMEAAGIHETTYNSIMKCDVDIRKDLYGNIVLSGG 304 Query: 887 STLFKGFGDRILSELKKLAPKDNKIRISAPQERLYSTWIGGSILASLDTFRRMWISKREF 1066 +T+F G DR+ E+ LAP KI++ AP ER YS WIGGSILASL TF++MWI+K E+ Sbjct: 305 TTMFPGIADRMSKEITALAPSSMKIKVVAPPERKYSVWIGGSILASLSTFQQMWIAKAEY 364 Query: 1067 EDDGYRALHRK 1099 ++ G +HRK Sbjct: 365 DESGPSIVHRK 375
>sp|P10365|ACT_THELA Actin Length = 375 Score = 425 bits (1092), Expect = e-118 Identities = 207/367 (56%), Positives = 272/367 (74%), Gaps = 4/367 (1%) Frame = +2 Query: 11 VVIDNGSGVIKAGFAGATTPKIHFPNYIGRPKHKRVMAGALEGDTFIGPKAEEHRGLMNI 190 +VIDNGSG+ KAGFAG P+ FP+ +GRP+H +M G + D+++G +A+ RG++++ Sbjct: 8 LVIDNGSGMCKAGFAGDDAPRAVFPSIVGRPRHHGIMIGMGQKDSYVGDEAQSKRGVLSL 67 Query: 191 RYPMEHGVVQDWNDMEKIWSYIYGPEQLKTSSEEHPVLLTEAPLNPLKNREKAAEIFFET 370 RYP+EHGVV +W+DMEKIW + + E L+ + EEHPVLLTEAP+NP NREK +I FET Sbjct: 68 RYPIEHGVVTNWDDMEKIWHHTFYNE-LRVAPEEHPVLLTEAPINPKSNREKMTQIVFET 126 Query: 371 FNVPALYISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFALQHAIMRTDIAGRDVS 550 FN PA Y+S+QAVLSLYA+GRTTG+VLDSGDGVTH VPIYEGFAL HAI R DIAGRD++ Sbjct: 127 FNAPAFYVSIQAVLSLYASGRTTGIVLDSGDGVTHTVPIYEGFALTHAISRIDIAGRDLT 186 Query: 551 RYLRLLLRKEGTHFQTSAEFEIVRQLKEKACFLSANPEMQEQSEIDNTT----YPLPDGS 718 YL +L + G F T+AE EIVR +KEK C+++ + E + Q+ ++T Y LPDG Sbjct: 187 DYLMKILAERGYSFTTTAEREIVRDIKEKLCYVAFDFEQELQTAAQSSTLEKSYELPDGQ 246 Query: 719 KLRIGGAKFRAPELLFRPDLIGSECDGIHEVLAYSIQKSDMDLRQILYGNIVLSGGSTLF 898 + IG +FRAPE LF P ++G E GIHE SI K D+D+R+ LYGNIV+SGG+T++ Sbjct: 247 VITIGNERFRAPEALFNPAVLGHESGGIHETTFNSIIKCDVDVRKDLYGNIVMSGGTTMY 306 Query: 899 KGFGDRILSELKKLAPKDNKIRISAPQERLYSTWIGGSILASLDTFRRMWISKREFEDDG 1078 G DR+ EL LAP KI+I AP ER YS WIGGSILASL TF++MW+SK+E+++ G Sbjct: 307 PGIADRMQKELTALAPSSMKIKIIAPPERKYSVWIGGSILASLSTFQQMWVSKQEYDESG 366 Query: 1079 YRALHRK 1099 +HRK Sbjct: 367 PSIVHRK 373
>sp|P53498|ACT_CHLRE Actin Length = 377 Score = 424 bits (1089), Expect = e-118 Identities = 202/367 (55%), Positives = 273/367 (74%), Gaps = 4/367 (1%) Frame = +2 Query: 11 VVIDNGSGVIKAGFAGATTPKIHFPNYIGRPKHKRVMAGALEGDTFIGPKAEEHRGLMNI 190 +V DNGSG++KAGFAG P+ FP+ +GRP+H VM G + D+++G +A+ RG++ + Sbjct: 10 LVCDNGSGMVKAGFAGDDAPRAVFPSIVGRPRHTGVMVGMGQKDSYVGDEAQSKRGILTL 69 Query: 191 RYPMEHGVVQDWNDMEKIWSYIYGPEQLKTSSEEHPVLLTEAPLNPLKNREKAAEIFFET 370 RYP+EHG+V +W+DMEKIW + + E L+ + EEHPVLLTEAPLNP NREK +I FET Sbjct: 70 RYPIEHGIVTNWDDMEKIWHHTFFNE-LRVAPEEHPVLLTEAPLNPKANREKMTQIMFET 128 Query: 371 FNVPALYISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFALQHAIMRTDIAGRDVS 550 FNVPA+Y+++QAVLSLYA+GRTTG+VLDSGDGVTH VPIYEG+AL HAI+R D+AGRD++ Sbjct: 129 FNVPAMYVAIQAVLSLYASGRTTGIVLDSGDGVTHTVPIYEGYALPHAILRLDLAGRDLT 188 Query: 551 RYLRLLLRKEGTHFQTSAEFEIVRQLKEKACFLSANPEMQEQSEIDNT----TYPLPDGS 718 YL +L + G F T+AE EIVR +KEK C+++ + E + + + ++ TY LPDG Sbjct: 189 DYLMKILMERGYSFTTTAEREIVRDIKEKLCYVALDFEQEMATALSSSALEKTYELPDGQ 248 Query: 719 KLRIGGAKFRAPELLFRPDLIGSECDGIHEVLAYSIQKSDMDLRQILYGNIVLSGGSTLF 898 + IG +FR PE+LF P++IG E GIH+ SI K D+D+R+ LY NIVLSGG+T+F Sbjct: 249 MITIGNERFRCPEVLFNPNMIGMEAVGIHDTTFNSIMKCDVDIRKDLYNNIVLSGGTTMF 308 Query: 899 KGFGDRILSELKKLAPKDNKIRISAPQERLYSTWIGGSILASLDTFRRMWISKREFEDDG 1078 G DR+ E+ LAP KI++ AP ER YS WIGGSILASL TF++MWI+K E+++ G Sbjct: 309 PGIADRMSKEITALAPSAMKIKVVAPPERKYSVWIGGSILASLSTFQQMWIAKSEYDESG 368 Query: 1079 YRALHRK 1099 +HRK Sbjct: 369 PSIVHRK 375
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 169,741,711
Number of Sequences: 369166
Number of extensions: 3821444
Number of successful extensions: 10857
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9559
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10159
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 15512822320
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail