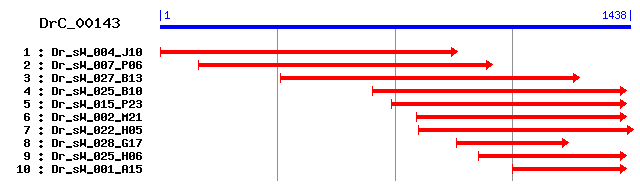
DrC_00143
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00143 (1437 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q26534|CATL_SCHMA Cathepsin L precursor (SMCL1) 318 3e-86 sp|Q9R013|CATF_MOUSE Cathepsin F precursor 317 4e-86 sp|Q9UBX1|CATF_HUMAN Cathepsin F precursor (CATSF) 309 1e-83 sp|Q9VN93|CPR1_DROME Putative cysteine proteinase CG12163 p... 290 7e-78 sp|P43296|RD19A_ARATH Cysteine proteinase RD19a precursor (... 261 3e-69 sp|P43295|A494_ARATH Probable cysteine proteinase A494 prec... 258 4e-68 sp|P04988|CYSP1_DICDI Cysteine proteinase 1 precursor 255 3e-67 sp|P25804|CYSP_PEA Cysteine proteinase 15A precursor (Turgo... 253 7e-67 sp|Q10716|CYSP1_MAIZE Cysteine proteinase 1 precursor 244 5e-64 sp|P14658|CYSP_TRYBB Cysteine proteinase precursor 243 1e-63
>sp|Q26534|CATL_SCHMA Cathepsin L precursor (SMCL1) Length = 319 Score = 318 bits (814), Expect = 3e-86 Identities = 154/300 (51%), Positives = 206/300 (68%), Gaps = 4/300 (1%) Frame = +1 Query: 475 FKKKFNKVYENDEEEEKRINLFHTNIKRSMVMRFLEKGTAEYGVTQFSDLTEEEFTNQYL 654 FK K+ K Y ++ E+E R N+F +NI ++ + + +G+A YGVT +SDLT +EF +L Sbjct: 23 FKLKYRKQY-HETEDEIRFNIFKSNILKAQLYQVFVRGSAIYGVTPYSDLTTDEFARTHL 81 Query: 655 SPKYDL----SKKPQRIAQIPQDIRTGQAIDWRVLGAVTPVKNQGSCGSCWAFSTTGNIE 822 + + + S P + + +I DWR GAVT VKNQG CGSCWAFSTTGN+E Sbjct: 82 TASWVVPSSRSNTPTSLGKEVNNIPKN--FDWREKGAVTEVKNQGMCGSCWAFSTTGNVE 139 Query: 823 GQWFIRTKRLVSLSEQQLVDCDTVDEGCNGGLPSQAYKVIQQMGGLETESDYPYKADRKT 1002 QWF +T +L+SLSEQQLVDCD +D+GCNGGLPS AY+ I +MGGL E +YPY A + Sbjct: 140 SQWFRKTGKLLSLSEQQLVDCDGLDDGCNGGLPSNAYESIIKMGGLMLEDNYPYDAKNEK 199 Query: 1003 CMLDKSKIAVYINGSESIDSSETTMAAWCSINGPISIGINAFAMQFYRGGISHPFKIFCN 1182 C L +AVYIN S ++ ET +AAW N IS+G+NA +QFY+ GISHP+ IFC+ Sbjct: 200 CHLKTDGVAVYINSSVNLTQDETELAAWLYHNSTISVGMNALLLQFYQHGISHPWWIFCS 259 Query: 1183 PDHLDHGVLIVGFNTTSSGEPFWIVKNSWGPGWGEDGYYRVFRGTGVCGLNKMPTSAIIH 1362 LDH VL+VG+ + EPFWIVKNSWG WGE+GY+R++RG G CG+N + TSA+I+ Sbjct: 260 KYLLDHAVLLVGYGVSEKNEPFWIVKNSWGVEWGENGYFRMYRGDGSCGINTVATSAMIY 319
>sp|Q9R013|CATF_MOUSE Cathepsin F precursor Length = 462 Score = 317 bits (813), Expect = 4e-86 Identities = 152/301 (50%), Positives = 203/301 (67%), Gaps = 2/301 (0%) Frame = +1 Query: 466 FDLFKKKFNKVYENDEEEEKRINLFHTNIKRSMVMRFLEKGTAEYGVTQFSDLTEEEFTN 645 F F +N+ YE+ EE + R+ +F N+ R+ ++ L++GTA+YG+T+FSDLTEEEF Sbjct: 165 FKDFMTTYNRTYESREEAQWRLTVFARNMIRAQKIQALDRGTAQYGITKFSDLTEEEFHT 224 Query: 646 QYLSPKYDLSKKPQRIAQIPQDIR--TGQAIDWRVLGAVTPVKNQGSCGSCWAFSTTGNI 819 YL+P L K+ R + I DWR GAVT VKNQG CGSCWAFS TGN+ Sbjct: 225 IYLNPL--LQKESGRKMSPAKSINDLAPPEWDWRKKGAVTEVKNQGMCGSCWAFSVTGNV 282 Query: 820 EGQWFIRTKRLVSLSEQQLVDCDTVDEGCNGGLPSQAYKVIQQMGGLETESDYPYKADRK 999 EGQWF+ L+SLSEQ+L+DCD VD+ C GGLPS AY I+ +GGLETE DY Y+ + Sbjct: 283 EGQWFLNRGTLLSLSEQELLDCDKVDKACLGGLPSNAYAAIKNLGGLETEDDYGYQGHVQ 342 Query: 1000 TCMLDKSKIAVYINGSESIDSSETTMAAWCSINGPISIGINAFAMQFYRGGISHPFKIFC 1179 TC VYIN S + +E +AAW + GPIS+ INAF MQFYR GI+HPF+ C Sbjct: 343 TCNFSAQMAKVYINDSVELSRNENKIAAWLAQKGPISVAINAFGMQFYRHGIAHPFRPLC 402 Query: 1180 NPDHLDHGVLIVGFNTTSSGEPFWIVKNSWGPGWGEDGYYRVFRGTGVCGLNKMPTSAII 1359 +P +DH VL+VG+ S+ P+W +KNSWG WGE+GYY ++RG+G CG+N M +SA++ Sbjct: 403 SPWFIDHAVLLVGYGNRSN-IPYWAIKNSWGSDWGEEGYYYLYRGSGACGVNTMASSAVV 461 Query: 1360 H 1362 + Sbjct: 462 N 462
>sp|Q9UBX1|CATF_HUMAN Cathepsin F precursor (CATSF) Length = 484 Score = 309 bits (792), Expect = 1e-83 Identities = 153/310 (49%), Positives = 204/310 (65%), Gaps = 3/310 (0%) Frame = +1 Query: 439 DGDIVSGSNFDLFKKKFNKVYENDEEEEKRINLFHTNIKRSMVMRFLEKGTAEYGVTQFS 618 D + S F F +N+ YE+ EE R+++F N+ R+ ++ L++GTA+YGVT+FS Sbjct: 178 DLPVKMASIFKNFVITYNRTYESKEEARWRLSVFVNNMVRAQKIQALDRGTAQYGVTKFS 237 Query: 619 DLTEEEFTNQYLSPKYDLSKKP---QRIAQIPQDIRTGQAIDWRVLGAVTPVKNQGSCGS 789 DLTEEEF YL+ L K+P + A+ D+ + DWR GAVT VK+QG CGS Sbjct: 238 DLTEEEFRTIYLNTL--LRKEPGNKMKQAKSVGDLAPPEW-DWRSKGAVTKVKDQGMCGS 294 Query: 790 CWAFSTTGNIEGQWFIRTKRLVSLSEQQLVDCDTVDEGCNGGLPSQAYKVIQQMGGLETE 969 CWAFS TGN+EGQWF+ L+SLSEQ+L+DCD +D+ C GGLPS AY I+ +GGLETE Sbjct: 295 CWAFSVTGNVEGQWFLNQGTLLSLSEQELLDCDKMDKACMGGLPSNAYSAIKNLGGLETE 354 Query: 970 SDYPYKADRKTCMLDKSKIAVYINGSESIDSSETTMAAWCSINGPISIGINAFAMQFYRG 1149 DY Y+ ++C K VYIN S + +E +AAW + GPIS+ INAF MQFYR Sbjct: 355 DDYSYQGHMQSCNFSAEKAKVYINDSVELSQNEQKLAAWLAKRGPISVAINAFGMQFYRH 414 Query: 1150 GISHPFKIFCNPDHLDHGVLIVGFNTTSSGEPFWIVKNSWGPGWGEDGYYRVFRGTGVCG 1329 GIS P + C+P +DH VL+VG+ S PFW +KNSWG WGE GYY + RG+G CG Sbjct: 415 GISRPLRPLCSPWLIDHAVLLVGYGNRSD-VPFWAIKNSWGTDWGEKGYYYLHRGSGACG 473 Query: 1330 LNKMPTSAII 1359 +N M +SA++ Sbjct: 474 VNTMASSAVV 483
>sp|Q9VN93|CPR1_DROME Putative cysteine proteinase CG12163 precursor Length = 614 Score = 290 bits (742), Expect = 7e-78 Identities = 143/312 (45%), Positives = 196/312 (62%), Gaps = 14/312 (4%) Frame = +1 Query: 466 FDLFKKKFNKVYENDEEEEKRINLFHTNIKRSMVMRFLEKGTAEYGVTQFSDLTEEEFTN 645 F F+ +F + Y + E + R+ +F N+K + E G+A+YG+T+F+D+T E+ Sbjct: 308 FYKFQVRFGRRYVSTAERQMRLRIFRQNLKTIEELNANEMGSAKYGITEFADMTSSEYKE 367 Query: 646 QYLSPKYDLSKK--------PQRIAQIPQDIRTGQAIDWRVLGAVTPVKNQGSCGSCWAF 801 + + D +K P ++P++ DWR AVT VKNQGSCGSCWAF Sbjct: 368 RTGLWQRDEAKATGGSAAVVPAYHGELPKEF------DWRQKDAVTQVKNQGSCGSCWAF 421 Query: 802 STTGNIEGQWFIRTKRLVSLSEQQLVDCDTVDEGCNGGLPSQAYKVIQQMGGLETESDYP 981 S TGNIEG + ++T L SEQ+L+DCDT D CNGGL AYK I+ +GGLE E++YP Sbjct: 422 SVTGNIEGLYAVKTGELKEFSEQELLDCDTTDSACNGGLMDNAYKAIKDIGGLEYEAEYP 481 Query: 982 YKADRKTCMLDKSKIAVYINGSESI-DSSETTMAAWCSINGPISIGINAFAMQFYRGGIS 1158 YKA + C +++ V + G + +ET M W NGPISIGINA AMQFYRGG+S Sbjct: 482 YKAKKNQCHFNRTLSHVQVAGFVDLPKGNETAMQEWLLANGPISIGINANAMQFYRGGVS 541 Query: 1159 HPFKIFCNPDHLDHGVLIVGFNTTSSGE-----PFWIVKNSWGPGWGEDGYYRVFRGTGV 1323 HP+K C+ +LDHGVL+VG+ + P+WIVKNSWGP WGE GYYRV+RG Sbjct: 542 HPWKALCSKKNLDHGVLVVGYGVSDYPNFHKTLPYWIVKNSWGPRWGEQGYYRVYRGDNT 601 Query: 1324 CGLNKMPTSAII 1359 CG+++M TSA++ Sbjct: 602 CGVSEMATSAVL 613
>sp|P43296|RD19A_ARATH Cysteine proteinase RD19a precursor (RD19) Length = 368 Score = 261 bits (668), Expect = 3e-69 Identities = 141/316 (44%), Positives = 195/316 (61%), Gaps = 18/316 (5%) Frame = +1 Query: 448 IVSGSNFDLFKKKFNKVYENDEEEEKRINLFHTNIKRSMVMRFLEKGTAEYGVTQFSDLT 627 + S +F LFK+KF KVY ++EE + R ++F N++R+ + L+ +A +GVTQFSDLT Sbjct: 45 LTSEDHFSLFKRKFGKVYASNEEHDYRFSVFKANLRRARRHQKLDP-SATHGVTQFSDLT 103 Query: 628 EEEFTNQYLSPK--YDLSKKPQRIAQIPQDIRTGQAIDWRVLGAVTPVKNQGSCGSCWAF 801 EF ++L + + L K + +P + + DWR GAVTPVKNQGSCGSCW+F Sbjct: 104 RSEFRKKHLGVRSGFKLPKDANKAPILPTE-NLPEDFDWRDHGAVTPVKNQGSCGSCWSF 162 Query: 802 STTGNIEGQWFIRTKRLVSLSEQQLVDC---------DTVDEGCNGGLPSQAYKVIQQMG 954 S TG +EG F+ T +LVSLSEQQLVDC D+ D GCNGGL + A++ + G Sbjct: 163 SATGALEGANFLATGKLVSLSEQQLVDCDHECDPEEADSCDSGCNGGLMNSAFEYTLKTG 222 Query: 955 GLETESDYPYKA-DRKTCMLDKSKIAVYINGSESIDSSETTMAAWCSINGPISIGINAFA 1131 GL E DYPY D KTC LDKSKI ++ I E +AA NGP+++ INA Sbjct: 223 GLMKEEDYPYTGKDGKTCKLDKSKIVASVSNFSVISIDEEQIAANLVKNGPLAVAINAGY 282 Query: 1132 MQFYRGGISHPFKIFCNPDHLDHGVLIVGFNTTS------SGEPFWIVKNSWGPGWGEDG 1293 MQ Y GG+S P+ C L+HGVL+VG+ +P+WI+KNSWG WGE+G Sbjct: 283 MQTYIGGVSCPY--ICT-RRLNHGVLLVGYGAAGYAPARFKEKPYWIIKNSWGETWGENG 339 Query: 1294 YYRVFRGTGVCGLNKM 1341 +Y++ +G +CG++ M Sbjct: 340 FYKICKGRNICGVDSM 355
>sp|P43295|A494_ARATH Probable cysteine proteinase A494 precursor Length = 361 Score = 258 bits (658), Expect = 4e-68 Identities = 139/314 (44%), Positives = 194/314 (61%), Gaps = 18/314 (5%) Frame = +1 Query: 454 SGSNFDLFKKKFNKVYENDEEEEKRINLFHTNIKRSMVMRFLEKGTAEYGVTQFSDLTEE 633 S +F LFKKKF KVY + EE R ++F N+ R+M + ++ +A +GVTQFSDLT Sbjct: 44 SEDHFTLFKKKFGKVYGSIEEHYYRFSVFKANLLRAMRHQKMDP-SARHGVTQFSDLTRS 102 Query: 634 EFTNQYLSPK--YDLSKKPQRIAQIPQDIRTGQAIDWRVLGAVTPVKNQGSCGSCWAFST 807 EF ++L K + L K + +P + DWR GAVTPVKNQGSCGSCW+FST Sbjct: 103 EFRRKHLGVKGGFKLPKDANQAPILPTQ-NLPEEFDWRDRGAVTPVKNQGSCGSCWSFST 161 Query: 808 TGNIEGQWFIRTKRLVSLSEQQLVDCD---------TVDEGCNGGLPSQAYKVIQQMGGL 960 TG +EG F+ T +LVSLSEQQLVDCD + D GCNGGL + A++ + GGL Sbjct: 162 TGALEGAHFLATGKLVSLSEQQLVDCDHECDPEEEGSCDSGCNGGLMNSAFEYTLKTGGL 221 Query: 961 ETESDYPYK-ADRKTCMLDKSKIAVYINGSESIDSSETTMAAWCSINGPISIGINAFAMQ 1137 E DYPY D +C LD+SKI ++ + +E +AA NGP+++ INA MQ Sbjct: 222 MREKDYPYTGTDGGSCKLDRSKIVASVSNFSVVSINEDQIAANLIKNGPLAVAINAAYMQ 281 Query: 1138 FYRGGISHPFKIFCNPDHLDHGVLIVGFNTTSSGE------PFWIVKNSWGPGWGEDGYY 1299 Y GG+S P+ C+ L+HGVL+VG+ + + P+WI+KNSWG WGE+G+Y Sbjct: 282 TYIGGVSCPY--ICS-RRLNHGVLLVGYGSAGFSQARLKEKPYWIIKNSWGESWGENGFY 338 Query: 1300 RVFRGTGVCGLNKM 1341 ++ +G +CG++ + Sbjct: 339 KICKGRNICGVDSL 352
>sp|P04988|CYSP1_DICDI Cysteine proteinase 1 precursor Length = 343 Score = 255 bits (651), Expect = 3e-67 Identities = 140/321 (43%), Positives = 187/321 (58%), Gaps = 21/321 (6%) Frame = +1 Query: 460 SNFDLFKKKFNKVYENDEEEEKRINLFHTNIKRSMVMRFL---EKGTAEYGVTQFSDLTE 630 S F F+ KFNK Y ++E E R +F +N+ + + + K ++GV +F+DL+ Sbjct: 27 SQFLEFQDKFNKKYSHEEYLE-RFEIFKSNLGKIEELNLIAINHKADTKFGVNKFADLSS 85 Query: 631 EEFTNQYLSPKYDLSKKPQRIAQIPQDIRTGQ---AIDWRVLGAVTPVKNQGSCGSCWAF 801 +EF N YL+ K + +A D A DWR GAVTPVKNQG CGSCW+F Sbjct: 86 DEFKNYYLNNKEAIFTDDLPVADYLDDEFINSIPTAFDWRTRGAVTPVKNQGQCGSCWSF 145 Query: 802 STTGNIEGQWFIRTKRLVSLSEQQLVDCD----------TVDEGCNGGLPSQAYKVIQQM 951 STTGN+EGQ FI +LVSLSEQ LVDCD DEGCNGGL AY I + Sbjct: 146 STTGNVEGQHFISQNKLVSLSEQNLVDCDHECMEYEGEEACDEGCNGGLQPNAYNYIIKN 205 Query: 952 GGLETESDYPYKADRKT-CMLDKSKIAVYINGSESIDSSETTMAAWCSINGPISIGINAF 1128 GG++TES YPY A+ T C + + I I+ I +ET MA + GP++I +A Sbjct: 206 GGIQTESSYPYTAETGTQCNFNSANIGAKISNFTMIPKNETVMAGYIVSTGPLAIAADAV 265 Query: 1129 AMQFYRGGISHPFKIFCNPDHLDHGVLIVGFNTTSS----GEPFWIVKNSWGPGWGEDGY 1296 QFY GG+ F I CNP+ LDHG+LIVG++ ++ P+WIVKNSWG WGE GY Sbjct: 266 EWQFYIGGV---FDIPCNPNSLDHGILIVGYSAKNTIFRKNMPYWIVKNSWGADWGEQGY 322 Query: 1297 YRVFRGTGVCGLNKMPTSAII 1359 + RG CG++ +++II Sbjct: 323 IYLRRGKNTCGVSNFVSTSII 343
>sp|P25804|CYSP_PEA Cysteine proteinase 15A precursor (Turgor-responsive protein 15A) Length = 363 Score = 253 bits (647), Expect = 7e-67 Identities = 136/310 (43%), Positives = 185/310 (59%), Gaps = 17/310 (5%) Frame = +1 Query: 463 NFDLFKKKFNKVYENDEEEEKRINLFHTNIKRSMVMRFLEKGTAEYGVTQFSDLTEEEFT 642 +F FK KF+K Y EE + R +F +N+ ++ + + + TAE+G+T+FSDLT EF Sbjct: 47 HFTSFKSKFSKSYATKEEHDYRFGVFKSNLIKAKLHQNRDP-TAEHGITKFSDLTASEFR 105 Query: 643 NQYLSPKYDLSKKPQRIAQIPQDIRTG--QAIDWRVLGAVTPVKNQGSCGSCWAFSTTGN 816 Q+L K L + P + P T + DWR GAVTPVK+QGSCGSCWAFSTTG Sbjct: 106 RQFLGLKKRL-RLPAHAQKAPILPTTNLPEDFDWREKGAVTPVKDQGSCGSCWAFSTTGA 164 Query: 817 IEGQWFIRTKRLVSLSEQQLVDCDTV---------DEGCNGGLPSQAYKVIQQMGGLETE 969 +EG ++ T +LVSLSEQQLVDCD V D GCNGGL + A++ + + GG+ E Sbjct: 165 LEGAHYLATGKLVSLSEQQLVDCDHVCDPEQAGSCDSGCNGGLMNNAFEYLLESGGVVQE 224 Query: 970 SDYPYKADRKTCMLDKSKIAVYINGSESIDSSETTMAAWCSINGPISIGINAFAMQFYRG 1149 DY Y +C DKSK+ ++ + E +AA NGP+++ INA MQ Y Sbjct: 225 KDYAYTGRDGSCKFDKSKVVASVSNFSVVTLDEDQIAANLVKNGPLAVAINAAWMQTYMS 284 Query: 1150 GISHPFKIFCNPDHLDHGVLIVGFNTTS------SGEPFWIVKNSWGPGWGEDGYYRVFR 1311 G+S P+ C LDHGVL+VGF + +P+WI+KNSWG WGE GYY++ R Sbjct: 285 GVSCPY--VCAKSRLDHGVLLVGFGKGAYAPIRLKEKPYWIIKNSWGQNWGEQGYYKICR 342 Query: 1312 GTGVCGLNKM 1341 G VCG++ M Sbjct: 343 GRNVCGVDSM 352
>sp|Q10716|CYSP1_MAIZE Cysteine proteinase 1 precursor Length = 371 Score = 244 bits (623), Expect = 5e-64 Identities = 136/334 (40%), Positives = 195/334 (58%), Gaps = 26/334 (7%) Frame = +1 Query: 439 DGDIVSGSNFDLFKKKFNKVYENDEEEEKRINLFHTNIKRSMVMRFLEKGTAEYGVTQFS 618 D ++ + S+F F ++F K Y++ +E R+++F N++R+ + L+ +AE+GVT+FS Sbjct: 39 DLELNAESHFLSFVQRFGKSYKDADEHAYRLSVFKDNLRRARRHQLLDP-SAEHGVTKFS 97 Query: 619 DLTEEEFTNQYLSPKY-------DLSKKPQRIAQIPQDIRTGQAIDWRVLGAVTPVKNQG 777 DLT EF YL + +L + +P D DWR GAV PVKNQG Sbjct: 98 DLTPAEFRRTYLGLRKSRRALLRELGESAHEAPVLPTD-GLPDDFDWRDHGAVGPVKNQG 156 Query: 778 SCGSCWAFSTTGNIEGQWFIRTKRLVSLSEQQLVDCD---------TVDEGCNGGLPSQA 930 SCGSCW+FS +G +EG ++ T +L LSEQQ VDCD + D GCNGGL + A Sbjct: 157 SCGSCWSFSASGALEGAHYLATGKLEVLSEQQFVDCDHECDSSEPDSCDSGCNGGLMTTA 216 Query: 931 YKVIQQMGGLETESDYPYKADRKTCMLDKSKIAVYINGSESIDSSETTMAAWCSINGPIS 1110 + +Q+ GGLE+E DYPY C DKSKI + + E ++A +GP++ Sbjct: 217 FSYLQKAGGLESEKDYPYTGSDGKCKFDKSKIVASVQNFSVVSVDEAQISANLIKHGPLA 276 Query: 1111 IGINAFAMQFYRGGISHPFKIFCNPDHLDHGVLIVGFNTTS------SGEPFWIVKNSWG 1272 IGINA MQ Y GG+S P+ C HLDHGVL+VG+ + +P+WI+KNSWG Sbjct: 277 IGINAAYMQTYIGGVSCPY--ICG-RHLDHGVLLVGYGASGFAPIRLKDKPYWIIKNSWG 333 Query: 1273 PGWGEDGYYRVFRGTGV---CGLNKM-PTSAIIH 1362 WGE+GYY++ RG+ V CG++ M T + +H Sbjct: 334 ENWGENGYYKICRGSNVRNKCGVDSMVSTVSAVH 367
>sp|P14658|CYSP_TRYBB Cysteine proteinase precursor Length = 450 Score = 243 bits (619), Expect = 1e-63 Identities = 130/306 (42%), Positives = 184/306 (60%), Gaps = 8/306 (2%) Frame = +1 Query: 466 FDLFKKKFNKVYENDEEEEKRINLFHTNIKRSMVMRFLEKGTAEYGVTQFSDLTEEEFTN 645 F FKKK+ KVY++ +EE R F N++++ + A +GVT FSD+T EEF Sbjct: 41 FAAFKKKYGKVYKDAKEEAFRFRAFEENMEQAKIQAAANP-YATFGVTPFSDMTREEFRA 99 Query: 646 QYLSPKYDLSKKPQRIAQIPQDIRTGQA---IDWRVLGAVTPVKNQGSCGSCWAFSTTGN 816 +Y + + +R+ + ++ TG+A +DWR GAVTPVK QG CGSCWAFST GN Sbjct: 100 RYRNGASYFAAAQKRLRKTV-NVTTGRAPAAVDWREKGAVTPVKVQGQCGSCWAFSTIGN 158 Query: 817 IEGQWFIRTKRLVSLSEQQLVDCDTVDEGCNGGLPSQAYK-VIQQMGG-LETESDYPY-- 984 IEGQW + LVSLSEQ LV CDT+D GCNGGL A+ ++ GG + TE+ YPY Sbjct: 159 IEGQWQVAGNPLVSLSEQMLVSCDTIDSGCNGGLMDNAFNWIVNSNGGNVFTEASYPYVS 218 Query: 985 -KADRKTCMLDKSKIAVYINGSESIDSSETTMAAWCSINGPISIGINAFAMQFYRGGISH 1161 ++ C ++ +I I + E +AA+ + NGP++I ++A + Y GGI Sbjct: 219 GNGEQPQCQMNGHEIGAAITDHVDLPQDEDAIAAYLAENGPLAIAVDAESFMDYNGGI-- 276 Query: 1162 PFKIFCNPDHLDHGVLIVGFNTTSSGEPFWIVKNSWGPGWGEDGYYRVFRGTGVCGLNKM 1341 C LDHGVL+VG+N +S P+WI+KNSW WGEDGY R+ +GT C +N+ Sbjct: 277 --LTSCTSKQLDHGVLLVGYN-DNSNPPYWIIKNSWSNMWGEDGYIRIEKGTNQCLMNQA 333 Query: 1342 PTSAII 1359 +SA++ Sbjct: 334 VSSAVV 339
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 156,128,688
Number of Sequences: 369166
Number of extensions: 3173049
Number of successful extensions: 9000
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8125
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8516
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 17215449160
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail