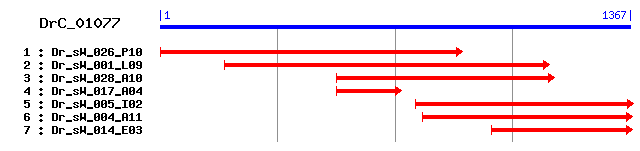
DrC_01077
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01077 (1367 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9DBI0|TMPS6_MOUSE Transmembrane protease, serine 6 (Mat... 141 4e-33 sp|P08419|ELA2_PIG Elastase-2 precursor 135 3e-31 sp|Q8IU80|TMPS6_HUMAN Transmembrane protease, serine 6 (Mat... 134 8e-31 sp|P05208|ELA2_MOUSE Elastase-2 precursor 130 7e-30 sp|P98074|ENTK_PIG Enteropeptidase precursor (Enterokinase)... 129 2e-29 sp|P26262|KLKB1_MOUSE Plasma kallikrein precursor (Plasma p... 127 8e-29 sp|P00774|ELA2_RAT Elastase-2 precursor 126 2e-28 sp|P14272|KLKB1_RAT Plasma kallikrein precursor (Plasma pre... 125 3e-28 sp|P03952|KLKB1_HUMAN Plasma kallikrein precursor (Plasma p... 124 5e-28 sp|P98072|ENTK_BOVIN Enteropeptidase precursor (Enterokinas... 124 5e-28
>sp|Q9DBI0|TMPS6_MOUSE Transmembrane protease, serine 6 (Matriptase-2) Length = 799 Score = 141 bits (356), Expect = 4e-33 Identities = 95/300 (31%), Positives = 148/300 (49%), Gaps = 6/300 (2%) Frame = +1 Query: 421 NCGIKPMEIENKIIGGEDSRKNAWPWHVGLYVNVRKRSIYICGGTLINPGWVVTAAHCLK 600 +CG++ + ++I+GG S + WPW L + R +ICGG LI WV+TAAHC + Sbjct: 555 DCGLQGLS--SRIVGGTVSSEGEWPWQASLQI----RGRHICGGALIADRWVITAAHCFQ 608 Query: 601 TIEAVN----SVPLGEMIDTRKYDLYDKFHVVINDHDRTTDDADEISRKLSYFIINPLFR 768 + +V LG+M ++ E+S K+S ++P Sbjct: 609 EDSMASPKLWTVFLGKMRQNSRW-------------------PGEVSFKVSRLFLHPYHE 649 Query: 769 ADYSSNLYDIALLKLDKPVIPSKVVNINFPCVPQVKNYTLDKSAKCYAIGWGNIEQNEPY 948 D S+ YD+ALL+LD PV+ S V P +++ + C+ GWG + P Sbjct: 650 ED--SHDYDVALLQLDHPVVYSATVR---PVCLPARSHFFEPGQHCWITGWGAQREGGPV 704 Query: 949 NIWSFEFDFVLERWRKNKSQKNSNILQEVEIPLVNFELCQKRYRIFNLNKEIHICAGSR- 1125 SN LQ+V++ LV +LC + YR + ++ + +CAG R Sbjct: 705 ----------------------SNTLQKVDVQLVPQDLCSEAYR-YQVSPRM-LCAGYRK 740 Query: 1126 -RKDTCRGDSGGGLFCLDPSTNRWQLYGITNFGSARGCGTSYGVYTRVPEVSNWIKEIIS 1302 +KD C+GDSGG L C +PS RW L G+ ++G G +GVYTRV V NWI+++++ Sbjct: 741 GKKDACQGDSGGPLVCREPS-GRWFLAGLVSWGLGCGRPNFFGVYTRVTRVINWIQQVLT 799
>sp|P08419|ELA2_PIG Elastase-2 precursor Length = 269 Score = 135 bits (340), Expect = 3e-31 Identities = 89/302 (29%), Positives = 146/302 (48%), Gaps = 5/302 (1%) Frame = +1 Query: 418 VNCGIKPMEIEN--KIIGGEDSRKNAWPWHVGLYVNVRKRSIYICGGTLINPGWVVTAAH 591 ++CG+ P + +++GGED+R N+WPW V L + + + CGGTL++ WV+TAAH Sbjct: 15 LSCGL-PANLPQLPRVVGGEDARPNSWPWQVSLQYDSSGQWRHTCGGTLVDQSWVLTAAH 73 Query: 592 CLKTIEAVNSVPLGEMIDTRKYDLYDKFHVVINDHDRTTDDADEISRKLSYFIINPLFRA 771 C+ + + VV+ H +T++ ++ K+S +++ + + Sbjct: 74 CISSSRT--------------------YRVVLGRHSLSTNEPGSLAVKVSKLVVHQDWNS 113 Query: 772 DYSSNLYDIALLKLDKPVIPSKVVNINFPCVPQVKNYTLDKSAKCYAIGWGNIEQNEPYN 951 + SN DIALLKL PV S I C+P L + CY GWG ++ N Sbjct: 114 NQLSNGNDIALLKLASPV--SLTDKIQLGCLP-AAGTILPNNYVCYVTGWGRLQTN---- 166 Query: 952 IWSFEFDFVLERWRKNKSQKNSNILQEVEIPLVNFELCQKRYRIFNLNKEIHICAGSRR- 1128 + +ILQ+ ++ +V++ C K + K ICAG Sbjct: 167 ------------------GASPDILQQGQLLVVDYATCSKPGWWGSTVKTNMICAGGDGI 208 Query: 1129 KDTCRGDSGGGLFCLDPSTNRWQLYGITNFGSARGCGTSY--GVYTRVPEVSNWIKEIIS 1302 +C GDSGG L C + +WQ++GI +FGS+ GC + V+TRV +WI +I+ Sbjct: 209 ISSCNGDSGGPLNC-QGANGQWQVHGIVSFGSSLGCNYYHKPSVFTRVSNYIDWINSVIA 267 Query: 1303 KS 1308 + Sbjct: 268 NN 269
>sp|Q8IU80|TMPS6_HUMAN Transmembrane protease, serine 6 (Matriptase-2) Length = 802 Score = 134 bits (336), Expect = 8e-31 Identities = 93/296 (31%), Positives = 148/296 (50%), Gaps = 2/296 (0%) Frame = +1 Query: 421 NCGIKPMEIENKIIGGEDSRKNAWPWHVGLYVNVRKRSIYICGGTLINPGWVVTAAHCLK 600 +CG++ ++I+GG S + WPW L V R +ICGG LI WV+TAAHC + Sbjct: 558 DCGLQGPS--SRIVGGAVSSEGEWPWQASLQVRGR----HICGGALIADRWVITAAHCFQ 611 Query: 601 TIEAVNSVPLGEMIDTRKYDLYDKFHVVINDHDRTTDDADEISRKLSYFIINPLFRADYS 780 ++V L+ F + + R E+S K+S +++P D Sbjct: 612 EDSMASTV------------LWTVFLGKVWQNSRWPG---EVSFKVSRLLLHPYHEED-- 654 Query: 781 SNLYDIALLKLDKPVIPSKVVNINFPCVPQVKNYTLDKSAKCYAIGWGNIEQNEPYNIWS 960 S+ YD+ALL+LD PV+ S V C+P +++ + C+ GWG + + P Sbjct: 655 SHDYDVALLQLDHPVVRSAAVRP--VCLP-ARSHFFEPGLHCWITGWGALREGGPI---- 707 Query: 961 FEFDFVLERWRKNKSQKNSNILQEVEIPLVNFELCQKRYRIFNLNKEIHICAGSRR--KD 1134 SN LQ+V++ L+ +LC + YR + + + +CAG R+ KD Sbjct: 708 ------------------SNALQKVDVQLIPQDLCSEVYR-YQVTPRM-LCAGYRKGKKD 747 Query: 1135 TCRGDSGGGLFCLDPSTNRWQLYGITNFGSARGCGTSYGVYTRVPEVSNWIKEIIS 1302 C+GDSGG L C S RW L G+ ++G G +GVYTR+ V +WI+++++ Sbjct: 748 ACQGDSGGPLVCKALS-GRWFLAGLVSWGLGCGRPNYFGVYTRITGVISWIQQVVT 802
>sp|P05208|ELA2_MOUSE Elastase-2 precursor Length = 271 Score = 130 bits (328), Expect = 7e-30 Identities = 85/303 (28%), Positives = 140/303 (46%), Gaps = 6/303 (1%) Frame = +1 Query: 418 VNCGIKPMEIEN---KIIGGEDSRKNAWPWHVGLYVNVRKRSIYICGGTLINPGWVVTAA 588 ++CG E+E+ +++GG+++ N WPW V L V R + CGG+L+ WV+TAA Sbjct: 15 LSCGYPTYEVEDDVSRVVGGQEATPNTWPWQVSLQVLSSGRWRHNCGGSLVANNWVLTAA 74 Query: 589 HCLKTIEAVNSVPLGEMIDTRKYDLYDKFHVVINDHDRTTDDADEISRKLSYFIINPLFR 768 HCL Y + V++ H + A + ++S +++ + Sbjct: 75 HCLSN--------------------YQTYRVLLGAHSLSNPGAGSAAVQVSKLVVHQRWN 114 Query: 769 ADYSSNLYDIALLKLDKPVIPSKVVNINFPCVPQVKNYTLDKSAKCYAIGWGNIEQNEPY 948 + N YDIAL+KL PV SK NI C+P L ++ CY GWG ++ N Sbjct: 115 SQNVGNGYDIALIKLASPVTLSK--NIQTACLPPA-GTILPRNYVCYVTGWGLLQTN--- 168 Query: 949 NIWSFEFDFVLERWRKNKSQKNSNILQEVEIPLVNFELCQKRYRIFNLNKEIHICAGSRR 1128 + + L++ + +V++ C + K +CAG Sbjct: 169 -------------------GNSPDTLRQGRLLVVDYATCSSASWWGSSVKSSMVCAGGDG 209 Query: 1129 -KDTCRGDSGGGLFCLDPSTNRWQLYGITNFGSARGCG--TSYGVYTRVPEVSNWIKEII 1299 +C GDSGG L C S +WQ++GI +FGS+ GC V+TRV +WI ++ Sbjct: 210 VTSSCNGDSGGPLNC-RASNGQWQVHGIVSFGSSLGCNYPRKPSVFTRVSNYIDWINSVM 268 Query: 1300 SKS 1308 +++ Sbjct: 269 ARN 271
>sp|P98074|ENTK_PIG Enteropeptidase precursor (Enterokinase) [Contains: Enteropeptidase non-catalytic mini chain; Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain] Length = 1034 Score = 129 bits (325), Expect = 2e-29 Identities = 91/298 (30%), Positives = 143/298 (47%), Gaps = 5/298 (1%) Frame = +1 Query: 421 NCGIKPM--EIENKIIGGEDSRKNAWPWHVGLYVNVRKRSIYICGGTLINPGWVVTAAHC 594 +CG K + E+ KI+GG DSR+ AWPW V LY N + +CG +L++ W+V+AAHC Sbjct: 786 SCGKKQVAQEVSPKIVGGNDSREGAWPWVVALYYNGQ----LLCGASLVSRDWLVSAAHC 841 Query: 595 LKTIEAVNSVPLGEMIDTRKYDLYDKFHVVINDHDRTTDDADEISRKLSYFIINPLFRAD 774 + G ++ K+ H+ N ++R + +INP + Sbjct: 842 V----------YGRNLEPSKWKAILGLHMTSN-----LTSPQIVTRLIDEIVINPHYNRR 886 Query: 775 YSSNLYDIALLKLDKPVIPSKVVNINFPCVPQVKNYTLDKSAKCYAIGWGN-IEQNEPYN 951 + DIA++ L+ V + I C+P+ +N C GWG I Q P Sbjct: 887 RKDS--DIAMMHLEFKV--NYTDYIQPICLPE-ENQVFPPGRICSIAGWGKVIYQGSP-- 939 Query: 952 IWSFEFDFVLERWRKNKSQKNSNILQEVEIPLVNFELCQKRYRIFNLNKEIHICAGSRRK 1131 ++ILQE ++PL++ E CQ++ +N+ + + +CAG Sbjct: 940 ---------------------ADILQEADVPLLSNEKCQQQMPEYNITENM-MCAGYEEG 977 Query: 1132 --DTCRGDSGGGLFCLDPSTNRWQLYGITNFGSARGCGTSYGVYTRVPEVSNWIKEII 1299 D+C+GDSGG L CL+ NRW L G+T+FG GVY RVP+ + WI+ + Sbjct: 978 GIDSCQGDSGGPLMCLE--NNRWLLAGVTSFGYQCALPNRPGVYARVPKFTEWIQSFL 1033
>sp|P26262|KLKB1_MOUSE Plasma kallikrein precursor (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain] Length = 638 Score = 127 bits (319), Expect = 8e-29 Identities = 92/287 (32%), Positives = 140/287 (48%), Gaps = 3/287 (1%) Frame = +1 Query: 442 EIENKIIGGEDSRKNAWPWHVGLYVNVRKRSIYICGGTLINPGWVVTAAHCLKTIEAVNS 621 +I +I+GG ++ WPW V L V + ++ ++CGG++I WV+TAAHC + Sbjct: 386 KINARIVGGTNASLGEWPWQVSLQVKLVSQT-HLCGGSIIGRQWVLTAAHCF------DG 438 Query: 622 VPLGEMIDTRKYDLYDKFHVVINDHDRTTDDADEISRKLSYFIINPLFRADYSSNLYDIA 801 +P D++ + +++ + T + S ++ II+ ++ S YDIA Sbjct: 439 IPYP--------DVWRIYGGILSLSEITKETP---SSRIKELIIHQEYKV--SEGNYDIA 485 Query: 802 LLKLDKPVIPSKVVNINFP-CVPQVKNYTLDKSAKCYAIGWGNIEQNEPYNIWSFEFDFV 978 L+KL P+ P C+P K T C+ GWG ++ Sbjct: 486 LIKLQTPL---NYTEFQKPICLPS-KADTNTIYTNCWVTGWGYTKEQG------------ 529 Query: 979 LERWRKNKSQKNSNILQEVEIPLVNFELCQKRYRIFNLNKEIHICAGSRR--KDTCRGDS 1152 + NILQ+ IPLV E CQK+YR + +NK++ ICAG + D C+GDS Sbjct: 530 ----------ETQNILQKATIPLVPNEECQKKYRDYVINKQM-ICAGYKEGGTDACKGDS 578 Query: 1153 GGGLFCLDPSTNRWQLYGITNFGSARGCGTSYGVYTRVPEVSNWIKE 1293 GG L C + RWQL GIT++G G GVYT+V E +WI E Sbjct: 579 GGPLVC--KHSGRWQLVGITSWGEGCGRKDQPGVYTKVSEYMDWILE 623
>sp|P00774|ELA2_RAT Elastase-2 precursor Length = 271 Score = 126 bits (316), Expect = 2e-28 Identities = 83/303 (27%), Positives = 141/303 (46%), Gaps = 6/303 (1%) Frame = +1 Query: 418 VNCGIKPMEIEN---KIIGGEDSRKNAWPWHVGLYVNVRKRSIYICGGTLINPGWVVTAA 588 ++CG E+++ +++GG+++ N+WPW V L + + CGG+L+ WV+TAA Sbjct: 15 LSCGYPTYEVQHDVSRVVGGQEASPNSWPWQVSLQYLSSGKWHHTCGGSLVANNWVLTAA 74 Query: 589 HCLKTIEAVNSVPLGEMIDTRKYDLYDKFHVVINDHDRTTDDADEISRKLSYFIINPLFR 768 HC+ ++R Y V++ H +T ++ ++ ++S +++ + Sbjct: 75 HCIS--------------NSRTY------RVLLGRHSLSTSESGSLAVQVSKLVVHEKWN 114 Query: 769 ADYSSNLYDIALLKLDKPVIPSKVVNINFPCVPQVKNYTLDKSAKCYAIGWGNIEQNEPY 948 A SN DIAL+KL PV + I C+P L + CY GWG ++ N Sbjct: 115 AQKLSNGNDIALVKLASPV--ALTSKIQTACLPPA-GTILPNNYPCYVTGWGRLQTN--- 168 Query: 949 NIWSFEFDFVLERWRKNKSQKNSNILQEVEIPLVNFELCQKRYRIFNLNKEIHICAGSRR 1128 ++LQ+ + +V++ C + K +CAG Sbjct: 169 -------------------GATPDVLQQGRLLVVDYATCSSASWWGSSVKTNMVCAGGDG 209 Query: 1129 -KDTCRGDSGGGLFCLDPSTNRWQLYGITNFGSARGCG--TSYGVYTRVPEVSNWIKEII 1299 +C GDSGG L C S +WQ++GI +FGS GC V+TRV +WI +I Sbjct: 210 VTSSCNGDSGGPLNC-QASNGQWQVHGIVSFGSTLGCNYPRKPSVFTRVSNYIDWINSVI 268 Query: 1300 SKS 1308 +K+ Sbjct: 269 AKN 271
>sp|P14272|KLKB1_RAT Plasma kallikrein precursor (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain] Length = 638 Score = 125 bits (314), Expect = 3e-28 Identities = 94/295 (31%), Positives = 137/295 (46%), Gaps = 6/295 (2%) Frame = +1 Query: 442 EIENKIIGGEDSRKNAWPWHVGLYVNVRKRSIYICGGTLINPGWVVTAAHCLKTIEAVNS 621 +I +I+GG +S WPW V L V + ++ ++CGG++I W++TAAHC + Sbjct: 386 KINARIVGGTNSSLGEWPWQVSLQVKLVSQN-HMCGGSIIGRQWILTAAHCF------DG 438 Query: 622 VPLGEMIDTRKYDLYDKFHVVINDHDRTTDDADEISRKLSYFIINPLF---RADYSSNLY 792 +P D++ + ++N EI+ K + I L + S Y Sbjct: 439 IPYP--------DVWRIYGGILN--------LSEITNKTPFSSIKELIIHQKYKMSEGSY 482 Query: 793 DIALLKLDKPVIPSKVVNINFP-CVPQVKNYTLDKSAKCYAIGWGNIEQNEPYNIWSFEF 969 DIAL+KL P+ P C+P K T C+ GWG ++ Sbjct: 483 DIALIKLQTPL---NYTEFQKPICLPS-KADTNTIYTNCWVTGWGYTKERG--------- 529 Query: 970 DFVLERWRKNKSQKNSNILQEVEIPLVNFELCQKRYRIFNLNKEIHICAGSRRK--DTCR 1143 + NILQ+ IPLV E CQK+YR + + K++ ICAG + D C+ Sbjct: 530 -------------ETQNILQKATIPLVPNEECQKKYRDYVITKQM-ICAGYKEGGIDACK 575 Query: 1144 GDSGGGLFCLDPSTNRWQLYGITNFGSARGCGTSYGVYTRVPEVSNWIKEIISKS 1308 GDSGG L C + RWQL GIT++G GVYT+V E +WI E I S Sbjct: 576 GDSGGPLVC--KHSGRWQLVGITSWGEGCARKEQPGVYTKVAEYIDWILEKIQSS 628
>sp|P03952|KLKB1_HUMAN Plasma kallikrein precursor (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain] Length = 638 Score = 124 bits (312), Expect = 5e-28 Identities = 93/283 (32%), Positives = 135/283 (47%), Gaps = 3/283 (1%) Frame = +1 Query: 454 KIIGGEDSRKNAWPWHVGLYVNVRKRSIYICGGTLINPGWVVTAAHCLKTIEAVNSVPLG 633 +I+GG +S WPW V L V + + ++CGG+LI WV+TAAHC + +PL Sbjct: 390 RIVGGTNSSWGEWPWQVSLQVKLTAQR-HLCGGSLIGHQWVLTAAHCF------DGLPL- 441 Query: 634 EMIDTRKYDLYDKFHVVINDHDRTTDDADEISRKLSYFIINPLFRADYSSNLYDIALLKL 813 D++ + ++N D T D ++ II+ ++ S +DIAL+KL Sbjct: 442 -------QDVWRIYSGILNLSDITKDTPFS---QIKEIIIHQNYKV--SEGNHDIALIKL 489 Query: 814 DKPVIPSKVVNINFP-CVPQVKNYTLDKSAKCYAIGWGNIEQNEPYNIWSFEFDFVLERW 990 P+ P C+P K T C+ GWG + Sbjct: 490 QAPL---NYTEFQKPICLPS-KGDTSTIYTNCWVTGWG---------------------F 524 Query: 991 RKNKSQKNSNILQEVEIPLVNFELCQKRYRIFNLNKEIHICAGSRR--KDTCRGDSGGGL 1164 K K + NILQ+V IPLV E CQKRY+ + + + + +CAG + KD C+GDSGG L Sbjct: 525 SKEKGEIQ-NILQKVNIPLVTNEECQKRYQDYKITQRM-VCAGYKEGGKDACKGDSGGPL 582 Query: 1165 FCLDPSTNRWQLYGITNFGSARGCGTSYGVYTRVPEVSNWIKE 1293 C W+L GIT++G GVYT+V E +WI E Sbjct: 583 VC--KHNGMWRLVGITSWGEGCARREQPGVYTKVAEYMDWILE 623
>sp|P98072|ENTK_BOVIN Enteropeptidase precursor (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain] Length = 1035 Score = 124 bits (312), Expect = 5e-28 Identities = 87/297 (29%), Positives = 143/297 (48%), Gaps = 4/297 (1%) Frame = +1 Query: 421 NCGIK--PMEIENKIIGGEDSRKNAWPWHVGLYVNVRKRSIYICGGTLINPGWVVTAAHC 594 +CG K E+ KI+GG DSR+ AWPW V LY + ++ +CG +L++ W+V+AAHC Sbjct: 787 SCGKKLVTQEVSPKIVGGSDSREGAWPWVVALYFDDQQ----VCGASLVSRDWLVSAAHC 842 Query: 595 LKTIEAVNSVPLGEMIDTRKYDLYDKFHVVINDHDRTTDDADEISRKLSYFIINPLFRAD 774 + G ++ K+ H+ N T+ + +R + +INP + Sbjct: 843 V----------YGRNMEPSKWKAVLGLHMASN---LTSPQIE--TRLIDQIVINPHYNKR 887 Query: 775 YSSNLYDIALLKLDKPVIPSKVVNINFPCVPQVKNYTLDKSAKCYAIGWGNIEQNEPYNI 954 +N DIA++ L+ V + I C+P+ +N C GWG + Sbjct: 888 RKNN--DIAMMHLEMKV--NYTDYIQPICLPE-ENQVFPPGRICSIAGWGALIY------ 936 Query: 955 WSFEFDFVLERWRKNKSQKNSNILQEVEIPLVNFELCQKRYRIFNLNKEIHICAG--SRR 1128 +++LQE ++PL++ E CQ++ +N+ + + +CAG + Sbjct: 937 ----------------QGSTADVLQEADVPLLSNEKCQQQMPEYNITENM-VCAGYEAGG 979 Query: 1129 KDTCRGDSGGGLFCLDPSTNRWQLYGITNFGSARGCGTSYGVYTRVPEVSNWIKEII 1299 D+C+GDSGG L C + NRW L G+T+FG GVY RVP + WI+ + Sbjct: 980 VDSCQGDSGGPLMCQE--NNRWLLAGVTSFGYQCALPNRPGVYARVPRFTEWIQSFL 1034
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 156,107,851
Number of Sequences: 369166
Number of extensions: 3420561
Number of successful extensions: 10822
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8901
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9684
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 16127659790
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail