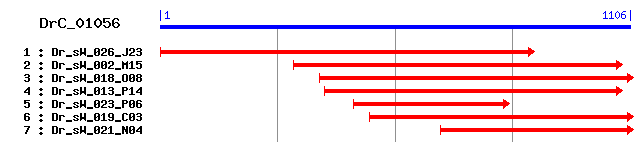
DrC_01056
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01056 (1105 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P53597|SUCA_HUMAN Succinyl-CoA ligase [GDP-forming] alph... 407 e-113 sp|Q9WUM5|SUCA_MOUSE Succinyl-CoA ligase [GDP-forming] alph... 398 e-110 sp|P13086|SUCA_RAT Succinyl-CoA ligase [GDP-forming] alpha-... 397 e-110 sp|Q94522|SUCA_DROME Succinyl-CoA ligase [GDP-forming] alph... 372 e-103 sp|P36967|SUCA_DICDI Succinyl-CoA ligase [GDP-forming] alph... 372 e-102 sp|P53596|SUCA_CAEEL Probable succinyl-CoA ligase [GDP-form... 360 5e-99 sp|O19069|SUCA_PIG Succinyl-CoA ligase [GDP-forming] alpha-... 354 3e-97 sp|Q8LAD2|SUCA2_ARATH Succinyl-CoA ligase [GDP-forming] alp... 340 4e-93 sp|P68209|SUCA1_ARATH Succinyl-CoA ligase [GDP-forming] alp... 340 4e-93 sp|O08371|SUCD_RICPR Succinyl-CoA synthetase alpha chain (S... 330 4e-90
>sp|P53597|SUCA_HUMAN Succinyl-CoA ligase [GDP-forming] alpha-chain, mitochondrial precursor (Succinyl-CoA synthetase, alpha chain) (SCS-alpha) Length = 333 Score = 407 bits (1047), Expect = e-113 Identities = 198/305 (64%), Positives = 236/305 (77%), Gaps = 1/305 (0%) Frame = +3 Query: 21 VRTC-YHRSRPHLSISRTTKVICQGLTGKQGTFHTKQAIDYGTHFVGGVSPGKGGQTHLD 197 +R C Y SR HL + + TK+ICQG TGKQGTFH++QA++YGT VGG +PGKGGQTHL Sbjct: 25 IRHCSYTASRQHLYVDKNTKIICQGFTGKQGTFHSQQALEYGTKLVGGTTPGKGGQTHLG 84 Query: 198 LPVFNTVKEAKEATNPDASVIFVPPPMAAASIMEAIDAEIPLVVCITEGIPQQDMVRVKN 377 LPVFNTVKEAKE T ASVI+VPPP AAA+I EAI+AEIPLVVCITEGIPQQDMVRVK+ Sbjct: 85 LPVFNTVKEAKEQTGATASVIYVPPPFAAAAINEAIEAEIPLVVCITEGIPQQDMVRVKH 144 Query: 378 RLINQEKTRLIGPNCPGIIKPNQCKIGIMPAHIHKPGKIGIVSRSGTLTYEAVHQTSQVG 557 +L+ QEKTRLIGPNCPG+I P +CKIGIMP HIHK G+IGIVSRSGTLTYEAVHQT+QVG Sbjct: 145 KLLRQEKTRLIGPNCPGVINPGECKIGIMPGHIHKKGRIGIVSRSGTLTYEAVHQTTQVG 204 Query: 558 LGQSLVIGIGGDPFNGTNFVDCLEVFVRDEHTKXXXXXXXXXXXXXXXXXMYLEKHNAGP 737 LGQSL +GIGGDPFNGT+F+DCLE+F+ D T+ +L++HN+GP Sbjct: 205 LGQSLCVGIGGDPFNGTDFIDCLEIFLNDSATEGIILIGEIGGNAEENAAEFLKQHNSGP 264 Query: 738 NRKAVVSFIAGLTAPPGRRMXXXXXXXXXXXXXXQEKIDALKHAGVIVTLSPAQMGASMI 917 N K VVSFIAGLTAPPGRRM +EKI AL+ AGV+V++SPAQ+G ++ Sbjct: 265 NSKPVVSFIAGLTAPPGRRMGHAGAIIAGGKGGAKEKISALQSAGVVVSMSPAQLGTTIY 324 Query: 918 EALHK 932 + K Sbjct: 325 KEFEK 329
>sp|Q9WUM5|SUCA_MOUSE Succinyl-CoA ligase [GDP-forming] alpha-chain, mitochondrial precursor (Succinyl-CoA synthetase, alpha chain) (SCS-alpha) Length = 333 Score = 398 bits (1022), Expect = e-110 Identities = 195/300 (65%), Positives = 228/300 (76%) Frame = +3 Query: 33 YHRSRPHLSISRTTKVICQGLTGKQGTFHTKQAIDYGTHFVGGVSPGKGGQTHLDLPVFN 212 Y SR H+ I + TK+ICQG TGKQGTFH++QA++YGT VGG +PGKGGQ HL LPVFN Sbjct: 30 YTASRKHIYIDKNTKIICQGFTGKQGTFHSQQALEYGTKLVGGTTPGKGGQKHLGLPVFN 89 Query: 213 TVKEAKEATNPDASVIFVPPPMAAASIMEAIDAEIPLVVCITEGIPQQDMVRVKNRLINQ 392 TVKEAKE T ASVI+VPPP AAA+I EAIDAEIPLVVCITEGIPQQDMVRVK+RL Q Sbjct: 90 TVKEAKEKTGATASVIYVPPPFAAAAINEAIDAEIPLVVCITEGIPQQDMVRVKHRLTRQ 149 Query: 393 EKTRLIGPNCPGIIKPNQCKIGIMPAHIHKPGKIGIVSRSGTLTYEAVHQTSQVGLGQSL 572 TRLIGPNCPG+I P +CKIGIMP HIHK G+IGIVSRSGTLTYEAVHQT+QVGLGQSL Sbjct: 150 GTTRLIGPNCPGVINPGECKIGIMPGHIHKKGRIGIVSRSGTLTYEAVHQTTQVGLGQSL 209 Query: 573 VIGIGGDPFNGTNFVDCLEVFVRDEHTKXXXXXXXXXXXXXXXXXMYLEKHNAGPNRKAV 752 IGIGGDPFNGT+F+DCLEVF+ D T+ +L++HN+GP K V Sbjct: 210 CIGIGGDPFNGTDFIDCLEVFLNDPATEGIILIGEIGGHAEENAAAFLKEHNSGPKAKPV 269 Query: 753 VSFIAGLTAPPGRRMXXXXXXXXXXXXXXQEKIDALKHAGVIVTLSPAQMGASMIEALHK 932 VSFIAG+TAPPGRRM +EKI AL+ AGV+V++SPAQ+G ++ + K Sbjct: 270 VSFIAGITAPPGRRMGHAGAIIAGGKGGAKEKISALQSAGVVVSMSPAQLGTTIYKEFEK 329
>sp|P13086|SUCA_RAT Succinyl-CoA ligase [GDP-forming] alpha-chain, mitochondrial precursor (Succinyl-CoA synthetase, alpha chain) (SCS-alpha) Length = 333 Score = 397 bits (1019), Expect = e-110 Identities = 196/300 (65%), Positives = 229/300 (76%) Frame = +3 Query: 33 YHRSRPHLSISRTTKVICQGLTGKQGTFHTKQAIDYGTHFVGGVSPGKGGQTHLDLPVFN 212 Y SR ++ I + TKVICQG TGKQGTFH++QA++YGT VGG +PGKGG+ HL LPVFN Sbjct: 30 YTASRKNIYIDKNTKVICQGFTGKQGTFHSQQALEYGTKLVGGTTPGKGGKKHLGLPVFN 89 Query: 213 TVKEAKEATNPDASVIFVPPPMAAASIMEAIDAEIPLVVCITEGIPQQDMVRVKNRLINQ 392 TVKEAKE T ASVI+VPPP AAA+I EAIDAEIPLVVCITEGIPQQDM+RVK++L Q Sbjct: 90 TVKEAKEKTGATASVIYVPPPFAAAAINEAIDAEIPLVVCITEGIPQQDMLRVKHKLTRQ 149 Query: 393 EKTRLIGPNCPGIIKPNQCKIGIMPAHIHKPGKIGIVSRSGTLTYEAVHQTSQVGLGQSL 572 KTRLIGPNCPGII P +CKIGIMP HIHK G+IGIVSRSGTLTYEAVHQT+QVGLGQSL Sbjct: 150 GKTRLIGPNCPGIINPGECKIGIMPGHIHKKGRIGIVSRSGTLTYEAVHQTTQVGLGQSL 209 Query: 573 VIGIGGDPFNGTNFVDCLEVFVRDEHTKXXXXXXXXXXXXXXXXXMYLEKHNAGPNRKAV 752 IGIGGDPFNGTNF+DCL+VF++D T+ +L++HN+GP K V Sbjct: 210 CIGIGGDPFNGTNFIDCLDVFLKDPATEGIVLIGEIGGHAEENAAEFLKEHNSGPKAKPV 269 Query: 753 VSFIAGLTAPPGRRMXXXXXXXXXXXXXXQEKIDALKHAGVIVTLSPAQMGASMIEALHK 932 VSFIAG+TAPPGRRM +EKI AL+ AGVIV++SPAQ+G M + K Sbjct: 270 VSFIAGITAPPGRRMGHAGAIIAGGKGGAKEKISALQSAGVIVSMSPAQLGTCMYKEFEK 329
>sp|Q94522|SUCA_DROME Succinyl-CoA ligase [GDP-forming] alpha-chain, mitochondrial precursor (Succinyl-CoA synthetase, alpha chain) (SCS-alpha) Length = 328 Score = 372 bits (956), Expect = e-103 Identities = 180/301 (59%), Positives = 230/301 (76%) Frame = +3 Query: 33 YHRSRPHLSISRTTKVICQGLTGKQGTFHTKQAIDYGTHFVGGVSPGKGGQTHLDLPVFN 212 Y+++R +L ++ ++VICQG TGKQGTFH++QA++YGT VGG+SP KGG HL LPVF Sbjct: 25 YNKTRGNLKLNGDSRVICQGFTGKQGTFHSQQALEYGTKLVGGISPKKGGTQHLGLPVFA 84 Query: 213 TVKEAKEATNPDASVIFVPPPMAAASIMEAIDAEIPLVVCITEGIPQQDMVRVKNRLINQ 392 +V EAK+AT+P A+VI+VPPP AAA+I+EA++AEIPL+VCITEG+PQ DMV+VK+ LI+Q Sbjct: 85 SVAEAKKATDPHATVIYVPPPGAAAAIIEALEAEIPLIVCITEGVPQHDMVKVKHALISQ 144 Query: 393 EKTRLIGPNCPGIIKPNQCKIGIMPAHIHKPGKIGIVSRSGTLTYEAVHQTSQVGLGQSL 572 K+RL+GPNCPGII P QCKIGIMP HIHK GKIG+VSRSGTLTYEAVHQT++VGLGQ+L Sbjct: 145 SKSRLVGPNCPGIIAPEQCKIGIMPGHIHKRGKIGVVSRSGTLTYEAVHQTTEVGLGQTL 204 Query: 573 VIGIGGDPFNGTNFVDCLEVFVRDEHTKXXXXXXXXXXXXXXXXXMYLEKHNAGPNRKAV 752 +GIGGDPFNGT+F+DCLEVF++D TK YL ++N+G K V Sbjct: 205 CVGIGGDPFNGTDFIDCLEVFLKDPETKGIILIGEIGGVAEEKAADYLTEYNSGIKAKPV 264 Query: 753 VSFIAGLTAPPGRRMXXXXXXXXXXXXXXQEKIDALKHAGVIVTLSPAQMGASMIEALHK 932 VSFIAG++APPGRRM +KI AL+ AGVIVT SPA+MG + + + + Sbjct: 265 VSFIAGVSAPPGRRMGHAGAIISGGKGGANDKIAALEKAGVIVTRSPAKMGHELFKEMKR 324 Query: 933 L 935 L Sbjct: 325 L 325
>sp|P36967|SUCA_DICDI Succinyl-CoA ligase [GDP-forming] alpha-chain, mitochondrial precursor (Succinyl-CoA synthetase, alpha chain) (SCS-alpha) Length = 310 Score = 372 bits (954), Expect = e-102 Identities = 189/295 (64%), Positives = 220/295 (74%) Frame = +3 Query: 33 YHRSRPHLSISRTTKVICQGLTGKQGTFHTKQAIDYGTHFVGGVSPGKGGQTHLDLPVFN 212 + ++P + I++ TKVICQG TG QGTFH+KQAI+YGT+ VGGVSPGKGGQ HLDLP FN Sbjct: 15 FFSTKPSVLINKNTKVICQGFTGNQGTFHSKQAIEYGTNMVGGVSPGKGGQKHLDLPAFN 74 Query: 213 TVKEAKEATNPDASVIFVPPPMAAASIMEAIDAEIPLVVCITEGIPQQDMVRVKNRLINQ 392 TVKEAKEAT +A+VI+VPPP AAA+I EAIDAE+ LVVCITEGIPQQDMV+VK L Q Sbjct: 75 TVKEAKEATGANATVIYVPPPHAAAAIKEAIDAEMELVVCITEGIPQQDMVKVKYLLNKQ 134 Query: 393 EKTRLIGPNCPGIIKPNQCKIGIMPAHIHKPGKIGIVSRSGTLTYEAVHQTSQVGLGQSL 572 KTRLIGPNCPGIIKP +CKIGIMP HIHKPGKIGIVSRSGTLTYEAV QT+ VGLGQS Sbjct: 135 NKTRLIGPNCPGIIKPGECKIGIMPGHIHKPGKIGIVSRSGTLTYEAVAQTTAVGLGQST 194 Query: 573 VIGIGGDPFNGTNFVDCLEVFVRDEHTKXXXXXXXXXXXXXXXXXMYLEKHNAGPNRKAV 752 IGIGGDPFNGTNF+DCL++F +D T+ +L + P K V Sbjct: 195 CIGIGGDPFNGTNFIDCLKMFTQDPQTEGIILIGEIGGEAEEEAAQWLIDN---PTDKPV 251 Query: 753 VSFIAGLTAPPGRRMXXXXXXXXXXXXXXQEKIDALKHAGVIVTLSPAQMGASMI 917 VSFIAGL+APPGRRM KI+ALK AGV VT SPA++G +++ Sbjct: 252 VSFIAGLSAPPGRRMGHAGAIISGGKGDANSKIEALKAAGVTVTFSPAKLGETIL 306
>sp|P53596|SUCA_CAEEL Probable succinyl-CoA ligase [GDP-forming] alpha-chain, mitochondrial precursor (Succinyl-CoA synthetase, alpha chain) (SCS-alpha) Length = 322 Score = 360 bits (923), Expect = 5e-99 Identities = 181/297 (60%), Positives = 213/297 (71%) Frame = +3 Query: 33 YHRSRPHLSISRTTKVICQGLTGKQGTFHTKQAIDYGTHFVGGVSPGKGGQTHLDLPVFN 212 Y+ + +L I++ TKVI QG TGKQGTFH KQ ++Y T VGGV+ K G HL LPVF Sbjct: 21 YNSTYNNLKINKDTKVIVQGFTGKQGTFHGKQMLEYNTKVVGGVNANKAGTEHLGLPVFK 80 Query: 213 TVKEAKEATNPDASVIFVPPPMAAASIMEAIDAEIPLVVCITEGIPQQDMVRVKNRLINQ 392 V EA+ T DASVI+VP A ++I EA+DAEIPLVVCITEGIPQ DMVRVK+RL+ Q Sbjct: 81 NVSEARNKTGADASVIYVPASAAGSAIEEAMDAEIPLVVCITEGIPQHDMVRVKSRLLKQ 140 Query: 393 EKTRLIGPNCPGIIKPNQCKIGIMPAHIHKPGKIGIVSRSGTLTYEAVHQTSQVGLGQSL 572 KTRL+GPNCPGII +QCKIGIMP HIHK G IGIVSRSGTLTYEAVHQT+QVG GQ+L Sbjct: 141 NKTRLVGPNCPGIISADQCKIGIMPGHIHKRGCIGIVSRSGTLTYEAVHQTTQVGFGQTL 200 Query: 573 VIGIGGDPFNGTNFVDCLEVFVRDEHTKXXXXXXXXXXXXXXXXXMYLEKHNAGPNRKAV 752 +GIGGDPFNGTNF+DCL VF+ D TK YL++HN+G NRK V Sbjct: 201 CVGIGGDPFNGTNFIDCLNVFLEDPETKGIILIGEIGGSAEEEAAAYLKEHNSGANRKPV 260 Query: 753 VSFIAGLTAPPGRRMXXXXXXXXXXXXXXQEKIDALKHAGVIVTLSPAQMGASMIEA 923 VSFIAG+TAPPGRRM +KI+AL+ AGV+VT SPA++G SM A Sbjct: 261 VSFIAGVTAPPGRRMGHAGAIISGGKGTAADKINALREAGVVVTDSPAKLGTSMATA 317
>sp|O19069|SUCA_PIG Succinyl-CoA ligase [GDP-forming] alpha-chain, mitochondrial precursor (Succinyl-CoA synthetase, alpha chain) (SCS-alpha) Length = 350 Score = 354 bits (908), Expect = 3e-97 Identities = 187/326 (57%), Positives = 226/326 (69%), Gaps = 22/326 (6%) Frame = +3 Query: 21 VRTC-YHRSRPHLSISRTTKVICQGLTGKQGTFHTKQAIDYGTHFVGGVSPGKGGQTHLD 197 +R C Y SR HL + + TKVICQG TGKQGTFH++QA++YGT+ VGG +PGKGG+THL Sbjct: 25 IRHCSYTASRKHLYVDKNTKVICQGFTGKQGTFHSQQALEYGTNLVGGTTPGKGGKTHLG 84 Query: 198 LPVFNTVKEAKEATNPDASVIFVPPPMAAASIMEAIDAEIPLVVCITEGIPQQDMVRVKN 377 LPVFNTVKEAKE T ASVI+VPPP AAA+I EAIDAE+PLVVCITEGIPQQDMVRVK+ Sbjct: 85 LPVFNTVKEAKEQTGATASVIYVPPPFAAAAINEAIDAEVPLVVCITEGIPQQDMVRVKH 144 Query: 378 RLINQEKTRLIGPNCPGII-------------------KPNQCKIGIM--PAHIHKPGKI 494 RL+ Q KTRLIGPNCPG+I + KI ++ IH + Sbjct: 145 RLLRQGKTRLIGPNCPGVINRFLRFAVYITSRVFTCTQQEEYRKIPLLFGTRSIH----M 200 Query: 495 GIVSRSGTLTYEAVHQTSQVGLGQSLVIGIGGDPFNGTNFVDCLEVFVRDEHTKXXXXXX 674 GIVSRSGTLTYEAVHQT+QVGLGQSL +GIGGDPFNGT+F DCLE+F+ D T+ Sbjct: 201 GIVSRSGTLTYEAVHQTTQVGLGQSLCVGIGGDPFNGTDFTDCLEIFLNDPATEGIILIG 260 Query: 675 XXXXXXXXXXXMYLEKHNAGPNRKAVVSFIAGLTAPPGRRMXXXXXXXXXXXXXXQEKID 854 +L++HN+GP K VVSFIAGLTAPPGRRM +EKI Sbjct: 261 EIGGNAEENAAEFLKQHNSGPKSKPVVSFIAGLTAPPGRRMGHAGAIIAGGKGGAKEKIT 320 Query: 855 ALKHAGVIVTLSPAQMGASMIEALHK 932 AL+ AGV+V++SPAQ+G ++ + K Sbjct: 321 ALQSAGVVVSMSPAQLGTTIYKEFEK 346
>sp|Q8LAD2|SUCA2_ARATH Succinyl-CoA ligase [GDP-forming] alpha-chain 2, mitochondrial precursor (Succinyl-CoA synthetase 2, alpha chain) (SCS2-alpha) Length = 341 Score = 340 bits (872), Expect = 4e-93 Identities = 175/287 (60%), Positives = 205/287 (71%) Frame = +3 Query: 60 ISRTTKVICQGLTGKQGTFHTKQAIDYGTHFVGGVSPGKGGQTHLDLPVFNTVKEAKEAT 239 + + T+VICQG+TGK GTFHT+QAI+YGT V GV+P KGG HL LPVFNTV EAK T Sbjct: 49 VDKNTRVICQGITGKNGTFHTEQAIEYGTKMVAGVTPKKGGTEHLGLPVFNTVAEAKAET 108 Query: 240 NPDASVIFVPPPMAAASIMEAIDAEIPLVVCITEGIPQQDMVRVKNRLINQEKTRLIGPN 419 +ASVI+VP P AAA+IME + AE+ L+VCITEGIPQ DMVRVK L +Q KTRLIGPN Sbjct: 109 KANASVIYVPAPFAAAAIMEGLAAELDLIVCITEGIPQHDMVRVKAALNSQSKTRLIGPN 168 Query: 420 CPGIIKPNQCKIGIMPAHIHKPGKIGIVSRSGTLTYEAVHQTSQVGLGQSLVIGIGGDPF 599 CPGIIKP +CKIGIMP +IHKPGKIGIVSRSGTLTYEAV QT+ VGLGQS +GIGGDPF Sbjct: 169 CPGIIKPGECKIGIMPGYIHKPGKIGIVSRSGTLTYEAVFQTTAVGLGQSTCVGIGGDPF 228 Query: 600 NGTNFVDCLEVFVRDEHTKXXXXXXXXXXXXXXXXXMYLEKHNAGPNRKAVVSFIAGLTA 779 NGTNFVDCLE F D T+ ++++ K VV+FIAGLTA Sbjct: 229 NGTNFVDCLEKFFVDPQTEGIVLIGEIGGTAEEDAAALIKENGTD---KPVVAFIAGLTA 285 Query: 780 PPGRRMXXXXXXXXXXXXXXQEKIDALKHAGVIVTLSPAQMGASMIE 920 PPGRRM Q+KI +L+ AGV V SPA++GA+M E Sbjct: 286 PPGRRMGHAGAIVSGGKGTAQDKIKSLRDAGVKVVESPAKIGAAMFE 332
>sp|P68209|SUCA1_ARATH Succinyl-CoA ligase [GDP-forming] alpha-chain 1, mitochondrial precursor (Succinyl-CoA synthetase 1, alpha chain) (SCS1-alpha) Length = 347 Score = 340 bits (872), Expect = 4e-93 Identities = 179/308 (58%), Positives = 213/308 (69%), Gaps = 4/308 (1%) Frame = +3 Query: 9 LKSFVRTCYHRSRPH----LSISRTTKVICQGLTGKQGTFHTKQAIDYGTHFVGGVSPGK 176 L S ++ S PH + + + T+V+CQG+TGK GTFHT+QAI+YGT V GV+P K Sbjct: 33 LTSLTQSRSFASDPHPPAAVFVDKNTRVLCQGITGKNGTFHTEQAIEYGTKMVAGVTPKK 92 Query: 177 GGQTHLDLPVFNTVKEAKEATNPDASVIFVPPPMAAASIMEAIDAEIPLVVCITEGIPQQ 356 GG HL LPVFN+V EAK T +ASVI+VP P AAA+IME I+AE+ L+VCITEGIPQ Sbjct: 93 GGTEHLGLPVFNSVAEAKADTKANASVIYVPAPFAAAAIMEGIEAELDLIVCITEGIPQH 152 Query: 357 DMVRVKNRLINQEKTRLIGPNCPGIIKPNQCKIGIMPAHIHKPGKIGIVSRSGTLTYEAV 536 DMVRVK+ L +Q KTRLIGPNCPGIIKP +CKIGIMP +IHKPGKIGIVSRSGTLTYEAV Sbjct: 153 DMVRVKHALNSQSKTRLIGPNCPGIIKPGECKIGIMPGYIHKPGKIGIVSRSGTLTYEAV 212 Query: 537 HQTSQVGLGQSLVIGIGGDPFNGTNFVDCLEVFVRDEHTKXXXXXXXXXXXXXXXXXMYL 716 QT+ VGLGQS +GIGGDPFNGTNFVDCLE F D T+ + Sbjct: 213 FQTTAVGLGQSTCVGIGGDPFNGTNFVDCLEKFFVDPQTEGIVLIGEIGGTAEEDAAALI 272 Query: 717 EKHNAGPNRKAVVSFIAGLTAPPGRRMXXXXXXXXXXXXXXQEKIDALKHAGVIVTLSPA 896 + A K VV+FIAGLTAPPGRRM Q+KI +L AGV V SPA Sbjct: 273 K---ASGTEKPVVAFIAGLTAPPGRRMGHAGAIVSGGKGTAQDKIKSLNDAGVKVVESPA 329 Query: 897 QMGASMIE 920 ++G++M E Sbjct: 330 KIGSAMYE 337
>sp|O08371|SUCD_RICPR Succinyl-CoA synthetase alpha chain (SCS-alpha) Length = 292 Score = 330 bits (846), Expect = 4e-90 Identities = 166/292 (56%), Positives = 211/292 (72%) Frame = +3 Query: 60 ISRTTKVICQGLTGKQGTFHTKQAIDYGTHFVGGVSPGKGGQTHLDLPVFNTVKEAKEAT 239 I++ TKVICQG TG QGTFH++QAI YGT+ VGGV+PGKGG THL+LPV+NTV EAK T Sbjct: 5 INKKTKVICQGFTGSQGTFHSEQAIAYGTNMVGGVTPGKGGHTHLNLPVYNTVHEAKAKT 64 Query: 240 NPDASVIFVPPPMAAASIMEAIDAEIPLVVCITEGIPQQDMVRVKNRLINQEKTRLIGPN 419 +ASVI+VPP AA SI+EAIDA+I +VVCITEGIP DM++VK LI KTRLIGPN Sbjct: 65 GANASVIYVPPGFAADSILEAIDAKIEVVVCITEGIPVLDMIKVKRALIG-SKTRLIGPN 123 Query: 420 CPGIIKPNQCKIGIMPAHIHKPGKIGIVSRSGTLTYEAVHQTSQVGLGQSLVIGIGGDPF 599 CPG+I P +CKIGIMP HIHK G IGIVSRSGTLTYEAV QT+ GLGQS +GIGGDP Sbjct: 124 CPGVITPGECKIGIMPGHIHKIGDIGIVSRSGTLTYEAVAQTTAAGLGQSTCVGIGGDPV 183 Query: 600 NGTNFVDCLEVFVRDEHTKXXXXXXXXXXXXXXXXXMYLEKHNAGPNRKAVVSFIAGLTA 779 NGT+FVDC+E+F++D+ TK ++++ +K +VSFIAG+TA Sbjct: 184 NGTSFVDCIEMFLQDDETKAIIMIGEIGGSAEEDAADFIKQSKI---KKPIVSFIAGITA 240 Query: 780 PPGRRMXXXXXXXXXXXXXXQEKIDALKHAGVIVTLSPAQMGASMIEALHKL 935 P +RM ++K++ L+ AGVI+T SPA +G +M++ L+K+ Sbjct: 241 PADKRMGHAGAIISGGKGSAEDKVEVLQSAGVIITRSPADIGKTMLDLLNKI 292
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 123,397,925
Number of Sequences: 369166
Number of extensions: 2553396
Number of successful extensions: 6224
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5938
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6153
length of database: 68,354,980
effective HSP length: 112
effective length of database: 47,664,660
effective search space used: 12154488300
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail