DrC_00938
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
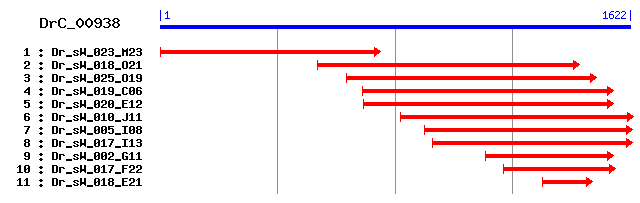
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00938 (1622 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q99NB8|UBQL4_MOUSE Ubiquilin-4 (Ataxin-1 ubiquitin-like ... 270 9e-72 sp|Q9NRR5|UBQL4_HUMAN Ubiquilin-4 (Ataxin-1 ubiquitin-like ... 270 1e-71 sp|Q8R317|UBQL1_MOUSE Ubiquilin-1 (Protein linking IAP with... 255 3e-67 sp|Q9UMX0|UBQL1_HUMAN Ubiquilin-1 (Protein linking IAP with... 250 7e-66 sp|Q9UHD9|UBQL2_HUMAN Ubiquilin-2 (Protein linking IAP with... 246 1e-64 sp|Q9JJP9|UBQL1_RAT Ubiquilin-1 (Protein linking IAP with c... 245 3e-64 sp|Q9QZM0|UBQL2_MOUSE Ubiquilin-2 (Protein linking IAP with... 230 1e-59 sp|Q8C5U9|UBQL3_MOUSE Ubiquilin-3 177 1e-43 sp|Q9H347|UBQL3_HUMAN Ubiquilin-3 164 7e-40 sp|P48510|DSK2_YEAST Ubiquitin-like protein DSK2 71 1e-11
>sp|Q99NB8|UBQL4_MOUSE Ubiquilin-4 (Ataxin-1 ubiquitin-like interacting protein A1U) Length = 596 Score = 270 bits (690), Expect = 9e-72 Identities = 189/543 (34%), Positives = 255/543 (46%), Gaps = 62/543 (11%) Frame = +3 Query: 3 IFAGKILKDEDKLASSKIVDGVTVHLVVKQLQQQTR-------------ASDVPLNQTNF 143 IFAGKILKD D L+ I DG+TVHLV+K Q+ ++ P Sbjct: 55 IFAGKILKDGDTLSQHGIKDGLTVHLVIKTPQKAQDPVTAAASPPSTPDSASAPSTTPAS 114 Query: 144 PGTVPVPPSQPTNTSSNIF--GQPMD----------------FAGLLGNMNTG---GSFD 260 P PV P NT+S+ G P F G+LG + G +F Sbjct: 115 PAAAPVQPCSSGNTTSDAGSGGGPSPVAAEGPSSATASILSGFGGILGLGSLGLGSANFM 174 Query: 261 DMQRQFQQQLMSNPDMMRSMLENPLMQQLLQSPEVIQTMMQSNPQVRQLIERNPEVGHIL 440 ++Q+Q Q+QLMSNP+M+ ++ENPL+Q ++ +P++++ M+ +NPQ++QL+ERNPE+ H+L Sbjct: 175 ELQQQMQRQLMSNPEMLSQIMENPLVQDMMSNPDLMRHMIMANPQMQQLMERNPEISHML 234 Query: 441 NNPSLMRQTMEMMRNPAMMQELMRHHDRALLNAEAFPGGMNHLTRLYRDVQEPLMDATT- 617 NNP LMRQTME+ RNPAMMQE+MR+ DRAL N E+ PGG N L R+Y D+QEP+ A Sbjct: 235 NNPELMRQTMELARNPAMMQEMMRNQDRALSNLESVPGGYNALRRMYTDIQEPMFTAARE 294 Query: 618 --GTNPFASLANRXXXXXXXRHGTENNEPLXXXXXXXXXXXXXXXXXXXXXXXXXXXGGN 791 G NPF+SLA TEN EPL G Sbjct: 295 QFGNNPFSSLAGNSDNSSSQPLRTENREPL----PNPWSPSPPTSQAPGSGGEGTGGSGT 350 Query: 792 TQINIPSNLPSAENLHNL-LGSFNDPRIQSLI-----------------------ETLRS 899 +Q++ + P N +L G FN P +Q+L+ +TL Sbjct: 351 SQVHPTVSNPFGINAASLGSGMFNSPEMQALLQQISENPQLMQNVISAPYMRTMMQTLAQ 410 Query: 900 NPQFTEMAIANSPA-AANPQLQEQLRNSMPQVLQAFERPEVRNALTNPRVLQAIFXXXXX 1076 NP F + N P A NPQLQEQLR +P LQ + PE + LTNPR +QA+ Sbjct: 411 NPDFAAQMMVNVPLFAGNPQLQEQLRLQLPVFLQQMQNPESLSILTNPRAMQALLQIQQG 470 Query: 1077 XXXXXXEAPHLXXXXXXXXXXXXXXXXXXXXXXXXXGGXXXXXXXXXXXXXXXXXXXXXX 1256 EAP L Sbjct: 471 LQTLQTEAPGLVPSLGSFGTPRTSVPLAGSNSGSSAEAPTSSPGVPATSPPSAGSNAQQQ 530 Query: 1257 XXXXXXXXNPGAGLNSAVGGNPEQLYSSELETMAQMGFPNRQANLRALIETFGDVNAAVN 1436 G+G NS V PE + +LE + MGF NR+ANL+ALI T GD+NAA+ Sbjct: 531 LMQQMIQLLSGSG-NSQV-PMPEVRFQQQLEQLNSMGFINREANLQALIATGGDINAAIE 588 Query: 1437 RIL 1445 R+L Sbjct: 589 RLL 591
>sp|Q9NRR5|UBQL4_HUMAN Ubiquilin-4 (Ataxin-1 ubiquitin-like interacting protein A1U) Length = 601 Score = 270 bits (689), Expect = 1e-71 Identities = 189/548 (34%), Positives = 256/548 (46%), Gaps = 67/548 (12%) Frame = +3 Query: 3 IFAGKILKDEDKLASSKIVDGVTVHLVVKQLQQQ------TRASDVPLNQTNFPGTVPVP 164 IFAGKILKD D L I DG+TVHLV+K Q+ T +S + + P T P Sbjct: 55 IFAGKILKDGDTLNQHGIKDGLTVHLVIKTPQKAQDPAAATASSPSTPDPASAPSTTPAS 114 Query: 165 PSQPTNTSSNIF-----------------------GQPM-------DFAGLLGNMNTG-- 248 P+ P S++ G P F G+LG + G Sbjct: 115 PATPAQPSTSGSASSDAGSGSRRSSGGGPSPGAGEGSPSATASILSGFGGILGLGSLGLG 174 Query: 249 -GSFDDMQRQFQQQLMSNPDMMRSMLENPLMQQLLQSPEVIQTMMQSNPQVRQLIERNPE 425 +F ++Q+Q Q+QLMSNP+M+ ++ENPL+Q ++ +P++++ M+ +NPQ++QL+ERNPE Sbjct: 175 SANFMELQQQMQRQLMSNPEMLSQIMENPLVQDMMSNPDLMRHMIMANPQMQQLMERNPE 234 Query: 426 VGHILNNPSLMRQTMEMMRNPAMMQELMRHHDRALLNAEAFPGGMNHLTRLYRDVQEPLM 605 + H+LNNP LMRQTME+ RNPAMMQE+MR+ DRAL N E+ PGG N L R+Y D+QEP+ Sbjct: 235 ISHMLNNPELMRQTMELARNPAMMQEMMRNQDRALSNLESIPGGYNALRRMYTDIQEPMF 294 Query: 606 DATT---GTNPFASLANRXXXXXXXRHGTENNEPLXXXXXXXXXXXXXXXXXXXXXXXXX 776 A G NPF+SLA TEN EPL Sbjct: 295 SAAREQFGNNPFSSLAGNSDSSSSQPLRTENREPL----PNPWSPSPPTSQAPGSGGEGT 350 Query: 777 XXGGNTQINIPSNLPSAENLHNL-LGSFNDPRIQ-----------------------SLI 884 G +Q++ + P N +L G FN P +Q S++ Sbjct: 351 GGSGTSQVHPTVSNPFGINAASLGSGMFNSPEMQALLQQISENPQLMQNVISAPYMRSMM 410 Query: 885 ETLRSNPQFTEMAIANSPA-AANPQLQEQLRNSMPQVLQAFERPEVRNALTNPRVLQAIF 1061 +TL NP F + N P A NPQLQEQLR +P LQ + PE + LTNPR +QA+ Sbjct: 411 QTLAQNPDFAAQMMVNVPLFAGNPQLQEQLRLQLPVFLQQMQNPESLSILTNPRAMQALL 470 Query: 1062 XXXXXXXXXXXEAPHLXXXXXXXXXXXXXXXXXXXXXXXXXGGXXXXXXXXXXXXXXXXX 1241 EAP L Sbjct: 471 QIQQGLQTLQTEAPGLVPSLGSFGISRTPAPSAGSNAGSTPEAPTSSPATPATSSPTGAS 530 Query: 1242 XXXXXXXXXXXXXNPGAGLNSAVGGNPEQLYSSELETMAQMGFPNRQANLRALIETFGDV 1421 G+G NS V PE + +LE + MGF NR+ANL+ALI T GD+ Sbjct: 531 SAQQQLMQQMIQLLAGSG-NSQV-QTPEVRFQQQLEQLNSMGFINREANLQALIATGGDI 588 Query: 1422 NAAVNRIL 1445 NAA+ R+L Sbjct: 589 NAAIERLL 596
>sp|Q8R317|UBQL1_MOUSE Ubiquilin-1 (Protein linking IAP with cytoskeleton-1) (PLIC-1) Length = 582 Score = 255 bits (651), Expect = 3e-67 Identities = 157/409 (38%), Positives = 222/409 (54%), Gaps = 40/409 (9%) Frame = +3 Query: 3 IFAGKILKDEDKLASSKIVDGVTVHLVVKQLQQQTRASDVPLNQTNFPGTV----PVPPS 170 IFAGKILKD+D L+ I DG+TVHLV+K Q R D QTN PG+ P P S Sbjct: 70 IFAGKILKDQDTLSQHGIHDGLTVHLVIKT---QNRPQDNSAQQTNAPGSTVTSSPAPDS 126 Query: 171 QPTNTSS--NIFGQPMDFAGLLGNMNTG---GSFDDMQRQFQQQLMSNPDMMRSMLENPL 335 PT+ S+ + FG GL G + G +F ++Q Q Q+QL+SNP+MM ++ENP Sbjct: 127 NPTSGSAANSSFGVG-GLGGLAGLSSLGLNTTNFSELQSQMQRQLLSNPEMMVQIMENPF 185 Query: 336 MQQLLQSPEVIQTMMQSNPQVRQLIERNPEVGHILNNPSLMRQTMEMMRNPAMMQELMRH 515 +Q +L +P++++ ++ +NPQ++QLI+RNPE+ H+LNNP +MRQT+E+ RNPAMMQE+MR+ Sbjct: 186 VQSMLSNPDLMRQLIMANPQMQQLIQRNPEISHMLNNPDIMRQTLELARNPAMMQEMMRN 245 Query: 516 HDRALLNAEAFPGGMNHLTRLYRDVQEPLMDATT---GTNPFASLANRXXXXXXXRHG-T 683 DRAL N E+ PGG N L R+Y D+QEP+++A G NPFASL + + T Sbjct: 246 QDRALSNLESIPGGYNALRRMYTDIQEPMLNAAQEQFGGNPFASLVSSSSSAEGTQPSRT 305 Query: 684 ENNEPLXXXXXXXXXXXXXXXXXXXXXXXXXXXGGNTQINIPS-NLPSAENLHNLLGS-- 854 EN +PL G T + P+ S +L G+ Sbjct: 306 ENRDPLPNPWAPQTSQSSPASGTTGSTTNTMSTSGGTATSTPAGQSTSGPSLVPGAGASM 365 Query: 855 FNDPRIQSLIETLRSNPQF------------------------TEMAIANSPAAANPQLQ 962 FN P +QSL++ + NPQ +M + N A NPQLQ Sbjct: 366 FNTPGMQSLLQQITENPQLMQNMLSAPYMRSMLQSLSQNPDLAAQMMLNNPLFAGNPQLQ 425 Query: 963 EQLRNSMPQVLQAFERPEVRNALTNPRVLQAIFXXXXXXXXXXXEAPHL 1109 EQ+R +P LQ + P+ +A++NPR +QA+ EAP L Sbjct: 426 EQMRQQLPTFLQQMQNPDTLSAMSNPRAMQALLQIQQGLQTLATEAPGL 474
Score = 53.5 bits (127), Expect = 2e-06
Identities = 27/56 (48%), Positives = 39/56 (69%)
Frame = +3
Query: 1290 AGLNSAVGGNPEQLYSSELETMAQMGFPNRQANLRALIETFGDVNAAVNRILQFDP 1457
AG+N + +PE + +LE ++ MGF NR+ANL+ALI T GD+NAA+ R+L P
Sbjct: 527 AGVNPQLQ-SPEVRFQQQLEQLSAMGFLNREANLQALIATGGDINAAIERLLGSQP 581
>sp|Q9UMX0|UBQL1_HUMAN Ubiquilin-1 (Protein linking IAP with cytoskeleton-1) (PLIC-1) (hPLIC-1) Length = 589 Score = 250 bits (639), Expect = 7e-66 Identities = 155/408 (37%), Positives = 217/408 (53%), Gaps = 39/408 (9%) Frame = +3 Query: 3 IFAGKILKDEDKLASSKIVDGVTVHLVVKQLQQQTRASDVPLNQTNFPGTVPVPPSQPTN 182 IFAGKILKD+D L+ I DG+TVHLV+K Q R D QTN G+ S P + Sbjct: 79 IFAGKILKDQDTLSQHGIHDGLTVHLVIKT---QNRPQDHSAQQTNTAGSNVTTSSTPNS 135 Query: 183 --TSSNIFGQPMDFAGLLG-------NMNTGGSFDDMQRQFQQQLMSNPDMMRSMLENPL 335 TS + P GL G +NT +F ++Q Q Q+QL+SNP+MM ++ENP Sbjct: 136 NSTSGSATSNPFGLGGLGGLAGLSSLGLNTT-NFSELQSQMQRQLLSNPEMMVQIMENPF 194 Query: 336 MQQLLQSPEVIQTMMQSNPQVRQLIERNPEVGHILNNPSLMRQTMEMMRNPAMMQELMRH 515 +Q +L +P++++ ++ +NPQ++QLI+RNPE+ H+LNNP +MRQT+E+ RNPAMMQE+MR+ Sbjct: 195 VQSMLSNPDLMRQLIMANPQMQQLIQRNPEISHMLNNPDIMRQTLELARNPAMMQEMMRN 254 Query: 516 HDRALLNAEAFPGGMNHLTRLYRDVQEPLMDATT---GTNPFASL-ANRXXXXXXXRHGT 683 DRAL N E+ PGG N L R+Y D+QEP++ A G NPFASL +N T Sbjct: 255 QDRALSNLESIPGGYNALRRMYTDIQEPMLSAAQEQFGGNPFASLVSNTSSGEGSQPSRT 314 Query: 684 ENNEPLXXXXXXXXXXXXXXXXXXXXXXXXXXXGGNTQINIPSNLPSAENLHNLLGS--F 857 EN +PL G+T +A NL +G+ F Sbjct: 315 ENRDPLPNPWAPQTSQSSSASSGTASTVGGTT--GSTASGTSGQSTTAPNLVPGVGASMF 372 Query: 858 NDPRIQSLIETLRSNPQF------------------------TEMAIANSPAAANPQLQE 965 N P +QSL++ + NPQ +M + N A NPQLQE Sbjct: 373 NTPGMQSLLQQITENPQLMQNMLSAPYMRSMMQSLSQNPDLAAQMMLNNPLFAGNPQLQE 432 Query: 966 QLRNSMPQVLQAFERPEVRNALTNPRVLQAIFXXXXXXXXXXXEAPHL 1109 Q+R +P LQ + P+ +A++NPR +QA+ EAP L Sbjct: 433 QMRQQLPTFLQQMQNPDTLSAMSNPRAMQALLQIQQGLQTLATEAPGL 480
Score = 55.5 bits (132), Expect = 4e-07
Identities = 28/56 (50%), Positives = 39/56 (69%)
Frame = +3
Query: 1290 AGLNSAVGGNPEQLYSSELETMAQMGFPNRQANLRALIETFGDVNAAVNRILQFDP 1457
AG+N + NPE + +LE ++ MGF NR+ANL+ALI T GD+NAA+ R+L P
Sbjct: 534 AGVNPQLQ-NPEVRFQQQLEQLSAMGFLNREANLQALIATGGDINAAIERLLGSQP 588
>sp|Q9UHD9|UBQL2_HUMAN Ubiquilin-2 (Protein linking IAP with cytoskeleton-2) (PLIC-2) (hPLIC-2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1) Length = 624 Score = 246 bits (628), Expect = 1e-64 Identities = 151/401 (37%), Positives = 216/401 (53%), Gaps = 32/401 (7%) Frame = +3 Query: 3 IFAGKILKDEDKLASSKIVDGVTVHLVVKQLQQQTRASDVPLNQTNFPGTVPVPP---SQ 173 IFAGKILKD+D L I DG+TVHLV+K + S P N T P S Sbjct: 75 IFAGKILKDQDTLIQHGIHDGLTVHLVIKSQNRPQGQSTQPSNAAGTNTTSASTPRSNST 134 Query: 174 PTNTSSNIFGQPMDFAGLLGNMNTGGS---FDDMQRQFQQQLMSNPDMMRSMLENPLMQQ 344 P +T+SN FG GL G + G S F ++Q Q QQQLM++P+MM ++ENP +Q Sbjct: 135 PISTNSNPFGLG-SLGGLAGLSSLGLSSTNFSELQSQMQQQLMASPEMMIQIMENPFVQS 193 Query: 345 LLQSPEVIQTMMQSNPQVRQLIERNPEVGHILNNPSLMRQTMEMMRNPAMMQELMRHHDR 524 +L +P++++ ++ +NPQ++QLI+RNPE+ H+LNNP +MRQT+E+ RNPAMMQE+MR+ D Sbjct: 194 MLSNPDLMRQLIMANPQMQQLIQRNPEISHLLNNPDIMRQTLEIARNPAMMQEMMRNQDL 253 Query: 525 ALLNAEAFPGGMNHLTRLYRDVQEPLMDATT---GTNPFASLANRXXXXXXXRHG-TENN 692 AL N E+ PGG N L R+Y D+QEP+++A G NPFAS+ + + TEN Sbjct: 254 ALSNLESIPGGYNALRRMYTDIQEPMLNAAQEQFGGNPFASVGSSSSSGEGTQPSRTENR 313 Query: 693 EPLXXXXXXXXXXXXXXXXXXXXXXXXXXXGGNTQINIPSNLPSAEN----------LHN 842 +PL GN+ N N +A N + + Sbjct: 314 DPL--PNPWAPPPATQSSATTSTTTSTGSGSGNSSSNATGNTVAAANYVASIFSTPGMQS 371 Query: 843 LLGSFND-----------PRIQSLIETLRSNPQFTEMAIANSPA-AANPQLQEQLRNSMP 986 LL + P ++S++++L NP + NSP ANPQLQEQ+R +P Sbjct: 372 LLQQITENPQLIQNMLSAPYMRSMMQSLSQNPDLAAQMMLNSPLFTANPQLQEQMRPQLP 431 Query: 987 QVLQAFERPEVRNALTNPRVLQAIFXXXXXXXXXXXEAPHL 1109 LQ + P+ +A++NPR +QA+ EAP L Sbjct: 432 AFLQQMQNPDTLSAMSNPRAMQALMQIQQGLQTLATEAPGL 472
Score = 56.2 bits (134), Expect = 3e-07
Identities = 28/56 (50%), Positives = 37/56 (66%)
Frame = +3
Query: 1290 AGLNSAVGGNPEQLYSSELETMAQMGFPNRQANLRALIETFGDVNAAVNRILQFDP 1457
AG N+ NPE + +LE + MGF NR+ANL+ALI T GD+NAA+ R+L P
Sbjct: 568 AGANAPQLPNPEVRFQQQLEQLNAMGFLNREANLQALIATGGDINAAIERLLGSQP 623
>sp|Q9JJP9|UBQL1_RAT Ubiquilin-1 (Protein linking IAP with cytoskeleton-1) (PLIC-1) Length = 582 Score = 245 bits (625), Expect = 3e-64 Identities = 152/409 (37%), Positives = 219/409 (53%), Gaps = 40/409 (9%) Frame = +3 Query: 3 IFAGKILKDEDKLASSKIVDGVTVHLVVKQLQQQTRASDVPLNQTNFPG----TVPVPPS 170 IFAGKILKD+D L+ I DG+TVHLV+K Q R D QTN G + P P S Sbjct: 70 IFAGKILKDQDTLSQHGIHDGLTVHLVIKT---QNRPQDNSAQQTNTTGNSVTSSPAPDS 126 Query: 171 QPTN--TSSNIFGQPMDFAGLLGNMNTG---GSFDDMQRQFQQQLMSNPDMMRSMLENPL 335 PT+ +++ FG GL G + G +F ++Q Q Q+QL+SNP+MM ++ENP Sbjct: 127 NPTSGPAANSSFGLG-GLGGLAGLSSLGLNTTNFSELQSQMQRQLLSNPEMMVQIMENPF 185 Query: 336 MQQLLQSPEVIQTMMQSNPQVRQLIERNPEVGHILNNPSLMRQTMEMMRNPAMMQELMRH 515 +Q +L +P++++ ++ +NPQ++QLI+RNPE+ H+LNNP++MRQT+E+ RNPAMMQE+MR+ Sbjct: 186 VQSMLSNPDLMRQLIMANPQMQQLIQRNPEISHMLNNPNIMRQTLELARNPAMMQEMMRN 245 Query: 516 HDRALLNAEAFPGGMNHLTRLYRDVQEPLMDATT---GTNPFASLANRXXXXXXXRHG-T 683 +R L N E+ PGG N L R+Y D+QEP+++A G NPFASL + + T Sbjct: 246 QERDLSNLESIPGGYNALRRMYTDIQEPMLNAAQEQFGGNPFASLVSSPSSAEGTQPSRT 305 Query: 684 ENNEPLXXXXXXXXXXXXXXXXXXXXXXXXXXXGGNTQINIPSNL-PSAENLHNLLGS-- 854 EN +PL + P+ S NL G+ Sbjct: 306 ENRDPLPNPWAPQTPQSSPASGSTGSTTNTVSTSAGNATSTPAGQGTSGPNLVPGAGASM 365 Query: 855 FNDPRIQSLIETLRSNPQF------------------------TEMAIANSPAAANPQLQ 962 FN P +QSL++ + NPQ +M + N A NPQLQ Sbjct: 366 FNTPGMQSLLQQITENPQLMQNMLSAPYMRSMMQSLSQNPDLAAQMMLNNPLFAGNPQLQ 425 Query: 963 EQLRNSMPQVLQAFERPEVRNALTNPRVLQAIFXXXXXXXXXXXEAPHL 1109 EQ+R +P LQ + P+ +A++NPR +QA+ EAP L Sbjct: 426 EQMRQQLPTFLQQMQNPDTLSAMSNPRAMQALLQIQQGLQTLATEAPGL 474
Score = 53.5 bits (127), Expect = 2e-06
Identities = 27/56 (48%), Positives = 39/56 (69%)
Frame = +3
Query: 1290 AGLNSAVGGNPEQLYSSELETMAQMGFPNRQANLRALIETFGDVNAAVNRILQFDP 1457
AG+N + +PE + +LE ++ MGF NR+ANL+ALI T GD+NAA+ R+L P
Sbjct: 527 AGVNPQLQ-SPEVRFQQQLEQLSAMGFLNREANLQALIATGGDINAAIERLLGSQP 581
>sp|Q9QZM0|UBQL2_MOUSE Ubiquilin-2 (Protein linking IAP with cytoskeleton-2) (PLIC-2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1) Length = 638 Score = 230 bits (586), Expect = 1e-59 Identities = 144/417 (34%), Positives = 218/417 (52%), Gaps = 48/417 (11%) Frame = +3 Query: 3 IFAGKILKDEDKLASSKIVDGVTVHLVVK-----QLQQQTRASDVPLNQTNFPGTV---- 155 IFAGKILKD+D L I DG+TVHLV+K Q Q T+ S T T Sbjct: 75 IFAGKILKDQDTLMQHGIHDGLTVHLVIKSQNRPQGQATTQPSTTAGTSTTTTTTTTAAA 134 Query: 156 --------PVPPSQPTNTSSNIFGQPMDFAGLLGNMNTGG----SFDDMQRQFQQQLMSN 299 P S PT T+S+ FG G L +++ G +F +++ Q QQQL+++ Sbjct: 135 PAATTSSAPRSSSTPTTTNSSSFG-----LGSLSSLSNLGLNSPNFTELRNQMQQQLLAS 189 Query: 300 PDMMRSMLENPLMQQLLQSPEVIQTMMQSNPQVRQLIERNPEVGHILNNPSLMRQTMEMM 479 P+MM ++ENP +Q +L +P++++ ++ +NPQ++QLI+RNPE+ H+LNNP +MRQT+E+ Sbjct: 190 PEMMIQIMENPFVQSMLSNPDLMRQLIMANPQMQQLIQRNPEISHLLNNPDIMRQTLEIA 249 Query: 480 RNPAMMQELMRHHDRALLNAEAFPGGMNHLTRLYRDVQEPLMDATT---GTNPFASLANR 650 RNPAMMQE+MR+ D AL N E+ PGG N L R+Y D+QEP+++A G NPFA++ + Sbjct: 250 RNPAMMQEMMRNQDLALSNLESIPGGYNALRRMYTDIQEPMLNAAQEQFGGNPFATVGSS 309 Query: 651 XXXXXXXRHG-TENNEPLXXXXXXXXXXXXXXXXXXXXXXXXXXXGGNTQ---------- 797 + TEN +PL G++ Sbjct: 310 STSGEGTQPSRTENRDPLPNPWAPPPTTQTAATTTTTTTTSSGSGSGSSSSSTTAGNTMA 369 Query: 798 -INIPSNLPSAENLHNLLGSFND-----------PRIQSLIETLRSNPQFTEMAIANSPA 941 N +++ S + +LL + P ++S++++L NP + +SP Sbjct: 370 AANYVASIFSTPGMQSLLQQITENPQLIQNMLSAPYMRSMMQSLSQNPDMAAQMMLSSPL 429 Query: 942 -AANPQLQEQLRNSMPQVLQAFERPEVRNALTNPRVLQAIFXXXXXXXXXXXEAPHL 1109 +NPQLQEQ+R +P LQ + PE A++NPR +QA+ EAP L Sbjct: 430 FTSNPQLQEQMRPQLPNFLQQMQNPETIAAMSNPRAMQALMQIQQGLQTLATEAPGL 486
Score = 53.5 bits (127), Expect = 2e-06
Identities = 25/47 (53%), Positives = 33/47 (70%)
Frame = +3
Query: 1317 NPEQLYSSELETMAQMGFPNRQANLRALIETFGDVNAAVNRILQFDP 1457
NPE + +LE + MGF NR+ANL+ALI T GD+NAA+ R+L P
Sbjct: 591 NPEVRFQQQLEQLNAMGFLNREANLQALIATGGDINAAIERLLGSQP 637
>sp|Q8C5U9|UBQL3_MOUSE Ubiquilin-3 Length = 658 Score = 177 bits (448), Expect = 1e-43 Identities = 98/242 (40%), Positives = 148/242 (61%), Gaps = 9/242 (3%) Frame = +3 Query: 3 IFAGKILKDEDKLASSKIVDGVTVHLVVKQLQQQTRASDVPLNQTNFPGTVPVPPSQPTN 182 IFAGKILKD D LA + DG+TVHLV+K +Q++T ++ P + PG P P P + Sbjct: 64 IFAGKILKDPDSLAQCGVRDGLTVHLVIK-MQRRTIGTECPSPPVSIPG--PNPGEIPQS 120 Query: 183 TSS-NIFGQPMDFAGLLGNMN----TGGSFDDMQRQFQQQLMSNPDMMRSMLENPLMQQL 347 +S ++ G P G+L ++ T GSF D Q +S P+++ ++++P +Q L Sbjct: 121 SSVYSVDGSPSFSLGVLTGLSGLGLTSGSFSDQPGSLMWQHISVPELVAQLVDDPFIQGL 180 Query: 348 LQSPEVIQTMMQSNPQVRQLIERNPEVGHILNNPSLMRQTMEMMRNPAMMQELMRHHDRA 527 L + +++ ++ NP ++ LI++NPE+GHILNNP +MRQTME +RNP+MMQE+MR DRA Sbjct: 181 LSNTGLVRQLVLDNPHMQHLIQQNPEIGHILNNPEIMRQTMEFLRNPSMMQEMMRSQDRA 240 Query: 528 LLNAEAFPGGMNHLTRLYRDVQEPLMDATT---GTNPFASLANRXXXXXXXRHG-TENNE 695 L N E+ PGG N L +Y D+ +P+++A G NPF + + TEN + Sbjct: 241 LSNLESIPGGYNVLRTMYTDIMDPMLNAVQEQFGGNPFVTATTASTTTTSSQPSRTENCD 300 Query: 696 PL 701 PL Sbjct: 301 PL 302
Score = 43.5 bits (101), Expect = 0.002
Identities = 24/54 (44%), Positives = 34/54 (62%), Gaps = 5/54 (9%)
Frame = +3
Query: 1296 LNSAVGGNPEQL-----YSSELETMAQMGFPNRQANLRALIETFGDVNAAVNRI 1442
L + NP+QL + +LE + MGF N +ANL+ALI T GDV+AAV ++
Sbjct: 602 LQALASTNPQQLQLEAHFRVQLEQLRAMGFLNLEANLQALIATEGDVDAAVEKL 655
>sp|Q9H347|UBQL3_HUMAN Ubiquilin-3 Length = 655 Score = 164 bits (415), Expect = 7e-40 Identities = 92/224 (41%), Positives = 136/224 (60%), Gaps = 12/224 (5%) Frame = +3 Query: 3 IFAGKILKDEDKLASSKIVDGVTVHLVVKQ----LQQQTRASDVPLNQTNFPGTVPVPPS 170 IFAGKILKD D LA + DG+TVHLV+K+ + + A+ VP Q PG++P P S Sbjct: 64 IFAGKILKDPDSLAQCGVRDGLTVHLVIKRQHRAMGNECPAASVP-TQGPSPGSLPQPSS 122 Query: 171 -QPTNTSSNIFGQPMDFAGLLGNMNTGG----SFDDMQRQFQQQLMSNPDMMRSMLENPL 335 P + G P GLL ++ G F D +Q +S P+ + ++++P Sbjct: 123 IYPAD------GPPAFSLGLLTGLSRLGLAYRGFPDQPSSLMRQHVSVPEFVTQLIDDPF 176 Query: 336 MQQLLQSPEVIQTMMQSNPQVRQLIERNPEVGHILNNPSLMRQTMEMMRNPAMMQELMRH 515 + LL + +++ ++ NP ++QLI+ NPE+GHILNNP +MRQT+E +RNPAMMQE++R Sbjct: 177 IPGLLSNTGLVRQLVLDNPHMQQLIQHNPEIGHILNNPEIMRQTLEFLRNPAMMQEMIRS 236 Query: 516 HDRALLNAEAFPGGMNHLTRLYRDVQEPLMDATT---GTNPFAS 638 DR L N E+ PGG N L +Y D+ +P+++A G NPFA+ Sbjct: 237 QDRVLSNLESIPGGYNVLCTMYTDIMDPMLNAVQEQFGGNPFAT 280
Score = 49.3 bits (116), Expect = 3e-05
Identities = 27/56 (48%), Positives = 36/56 (64%), Gaps = 5/56 (8%)
Frame = +3
Query: 1296 LNSAVGGNPEQL-----YSSELETMAQMGFPNRQANLRALIETFGDVNAAVNRILQ 1448
L V NP+QL + +LE + MGF NR+ANL+ALI T GDV+AAV ++ Q
Sbjct: 599 LQDLVSTNPQQLQPEAHFQVQLEQLRSMGFLNREANLQALIATGGDVDAAVEKLRQ 654
>sp|P48510|DSK2_YEAST Ubiquitin-like protein DSK2 Length = 373 Score = 70.9 bits (172), Expect = 1e-11 Identities = 75/248 (30%), Positives = 109/248 (43%), Gaps = 33/248 (13%) Frame = +3 Query: 3 IFAGKILKDEDKLASSKIVDGVTVHLVVKQLQQQTRASDVPLNQTNFPGTVPVPPSQPTN 182 I++GKILKD+ + S I DG +VHLV Q + QT AS N G + P Sbjct: 45 IYSGKILKDDQTVESYHIQDGHSVHLVKSQPKPQT-ASAAGANNATATGAAAGTGATPNM 103 Query: 183 TSSNIFG-QPM------DFAGLLGNMNTGGSFDDMQRQFQQQLMSNPDMMRSMLENPL-- 335 +S G P+ +AG L NM + F +N D + M+ENP+ Sbjct: 104 SSGQSRGFNPLADLTSARYAGYL-NMPSADMFGPDGGALNND-SNNQDELLRMMENPIFQ 161 Query: 336 --MQQLLQSPEVIQTMMQSNPQV-------RQLIERNPEVGHILNNPSLMRQTMEMMRNP 488 M ++L +P+++ M+QSNPQ+ RQ+++ +P +L NP ++RQ+M+ R Sbjct: 162 SQMNEMLSNPQMLDFMIQSNPQLQAMGPQARQMLQ-SPMFRQMLTNPDMIRQSMQFAR-- 218 Query: 489 AMMQELMRHHDRALLNAEAFPG--------GMNHLTRLYRDVQEPL-------MDATTGT 623 M + A A AFP G N T + A T Sbjct: 219 --MMDPNAGMGSAGGAASAFPAPGGDAPEEGSNTNTTSSSNTGNNAGTNAGTNAGANTAA 276 Query: 624 NPFASLAN 647 NPFASL N Sbjct: 277 NPFASLLN 284
Score = 37.7 bits (86), Expect = 0.097
Identities = 23/76 (30%), Positives = 35/76 (46%)
Frame = +3
Query: 825 AENLHNLLGSFNDPRIQSLIETLRSNPQFTEMAIANSPAAANPQLQEQLRNSMPQVLQAF 1004
+ N LL +P QS + + SNPQ + I ++P QL+ PQ Q
Sbjct: 145 SNNQDELLRMMENPIFQSQMNEMLSNPQMLDFMIQSNP---------QLQAMGPQARQML 195
Query: 1005 ERPEVRNALTNPRVLQ 1052
+ P R LTNP +++
Sbjct: 196 QSPMFRQMLTNPDMIR 211
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 180,325,905
Number of Sequences: 369166
Number of extensions: 3731022
Number of successful extensions: 10071
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9118
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9888
length of database: 68,354,980
effective HSP length: 116
effective length of database: 46,925,720
effective search space used: 19896505280
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail