DrC_00783
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00783 (1711 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P50990|TCPQ_HUMAN T-complex protein 1, theta subunit (TC... 561 e-159 sp|P42932|TCPQ_MOUSE T-complex protein 1, theta subunit (TC... 558 e-158 sp|P78921|TCPQ_SCHPO Probable T-complex protein 1, theta su... 435 e-121 sp|P47079|TCPQ_YEAST T-complex protein 1, theta subunit (TC... 402 e-111 sp|P47828|TCPQ_CANAL T-complex protein 1, theta subunit (TC... 345 3e-94 sp|Q58405|THS_METJA Thermosome subunit (Chaperonin subunit) 261 4e-69 sp|O93624|THS_METTL Thermosome subunit (Chaperonin subunit) 256 1e-67 sp|O26885|THSB_METTH Thermosome beta subunit (Thermosome su... 256 2e-67 sp|O28045|THSA_ARCFU Thermosome alpha subunit (Thermosome s... 255 2e-67 sp|P50016|THS_METKA Thermosome subunit (Chaperonin-like com... 249 2e-65
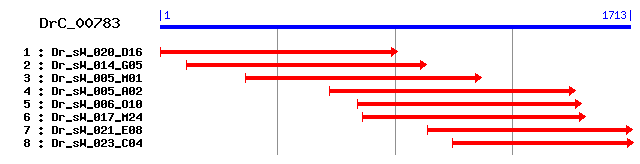
>sp|P50990|TCPQ_HUMAN T-complex protein 1, theta subunit (TCP-1-theta) (CCT-theta) sp|Q5RAP1|TCPQ_PONPY T-complex protein 1, theta subunit (TCP-1-theta) (CCT-theta) Length = 548 Score = 561 bits (1445), Expect = e-159 Identities = 276/525 (52%), Positives = 383/525 (72%) Frame = +1 Query: 1 AMHIPTAPGYLSMMKEGSRFYQGLEESVYRNIQACVALGQTTASTFGPQGMKKMVINHIG 180 A+H+P APG+ M+KEG++ + GLEE+VYRNIQAC L QTT + +GP GM KMVINH+ Sbjct: 2 ALHVPKAPGFAQMLKEGAKHFSGLEEAVYRNIQACKELAQTTRTAYGPNGMNKMVINHLE 61 Query: 181 KKFVTKDASTILRELEVEHPAAKLMVMXXXXXXXXSGEATNFVIILCASLLEKAEYLLKM 360 K FVT DA+TILRELEV+HPAAK++VM G+ TNFV++ +LLE AE LL++ Sbjct: 62 KLFVTNDAATILRELEVQHPAAKMIVMASHMQEQEVGDGTNFVLVFAGALLELAEELLRI 121 Query: 361 GLSVPDVTEGFEIAAKHSLEILKDLVIGKVEDLREKEMIKKIVGTAVNSKIFGEESHISD 540 GLSV +V EG+EIA + + EIL +LV ++LR+ + + ++ T++ SK +G E ++ Sbjct: 122 GLSVSEVIEGYEIACRKAHEILPNLVCCSAKNLRDIDEVSSLLRTSIMSKQYGNEVFLAK 181 Query: 541 LVVDACIRALPTSSFFNVDNIRVVKILGSGTNASKVFNGMVFVREAQSVIKECKEAKVVV 720 L+ AC+ P S FNVDNIRV KILGSG ++S V +GMVF +E + + K+AK+ V Sbjct: 182 LIAQACVSIFPDSGHFNVDNIRVCKILGSGISSSSVLHGMVFKKETEGDVTSVKDAKIAV 241 Query: 721 FSCPFDQLQTETKGTVLINTAKELMDFSRGEEDQMEERVREIAETGCKVIVTGGKVGELA 900 +SCPFD + TETKGTVLI TA+ELM+FS+GEE+ M+ +V+ IA+TG V+VTGGKV ++A Sbjct: 242 YSCPFDGMITETKGTVLIKTAEELMNFSKGEENLMDAQVKAIADTGANVVVTGGKVADMA 301 Query: 901 LHYANKYNLMVVKLSSKFDIRRVCKVTGATPIPKLSTPTQEELGYIASVRTEEIGKSTVV 1080 LHYANKYN+M+V+L+SK+D+RR+CK GAT +P+L+ P EE+G+ SV E+G + VV Sbjct: 302 LHYANKYNIMLVRLNSKWDLRRLCKTVGATALPRLTPPVLEEMGHCDSVYLSEVGDTQVV 361 Query: 1081 LFDQDVKEGAISSIVVRGSMYDMMDEVERAIDDGVNTFKCLTKEPRYLAGAGATEIELAN 1260 +F + ++GAIS+IV+RGS ++MD++ERA+DDGVNTFK LT++ R + G GATEIELA Sbjct: 362 VFKHEKEDGAISTIVLRGSTDNLMDDIERAVDDGVNTFKVLTRDKRLVPGGGATEIELAK 421 Query: 1261 RLSDFANTQTGLVQYAIKQFVLAFESIVKVLCDNAGENATERIAELKAHHREGKTTFGII 1440 +++ + T GL QYAIK+F AFE+I + L +N+G A E I++L A H+EG G+ Sbjct: 422 QITSYGETCPGLEQYAIKKFAEAFEAIPRALAENSGVKANEVISKLYAVHQEGNKNVGLD 481 Query: 1441 STSEGRKVDDAVEHQILDTYTAKYWGIRFAADTAVTILRVDNIIM 1575 +E V D +E ILDTY KYW I+ A + AVT+LRVD IIM Sbjct: 482 IEAEVPAVKDMLEAGILDTYLGKYWAIKLATNAAVTVLRVDQIIM 526
>sp|P42932|TCPQ_MOUSE T-complex protein 1, theta subunit (TCP-1-theta) (CCT-theta) Length = 548 Score = 558 bits (1438), Expect = e-158 Identities = 276/525 (52%), Positives = 381/525 (72%) Frame = +1 Query: 1 AMHIPTAPGYLSMMKEGSRFYQGLEESVYRNIQACVALGQTTASTFGPQGMKKMVINHIG 180 A+H+P APG+ M+K+G++ + GLEE+VYRNIQAC L QTT + +GP GM KMVIN + Sbjct: 2 ALHVPKAPGFAQMLKDGAKHFSGLEEAVYRNIQACKELAQTTRTAYGPNGMNKMVINRLE 61 Query: 181 KKFVTKDASTILRELEVEHPAAKLMVMXXXXXXXXSGEATNFVIILCASLLEKAEYLLKM 360 K FVT DA+TILRELEV+HPAAK++VM G+ TNFV++ +LLE AE LL++ Sbjct: 62 KLFVTNDAATILRELEVQHPAAKMIVMASHMQEQEVGDGTNFVLVFAGALLELAEELLRI 121 Query: 361 GLSVPDVTEGFEIAAKHSLEILKDLVIGKVEDLREKEMIKKIVGTAVNSKIFGEESHISD 540 GLSV +V G+EIA K + EIL +LV ++LR+ + + ++ T++ SK +G E+ ++ Sbjct: 122 GLSVSEVISGYEIACKKAHEILPELVCCSAKNLRDVDEVSSLLRTSIMSKQYGSETFLAK 181 Query: 541 LVVDACIRALPTSSFFNVDNIRVVKILGSGTNASKVFNGMVFVREAQSVIKECKEAKVVV 720 L+ AC+ P S FNVDNIRV KILGSG +S V +GMVF +E + + K+AK+ V Sbjct: 182 LIAQACVSIFPDSGNFNVDNIRVCKILGSGIYSSSVLHGMVFKKETEGDVTSVKDAKIAV 241 Query: 721 FSCPFDQLQTETKGTVLINTAKELMDFSRGEEDQMEERVREIAETGCKVIVTGGKVGELA 900 +SCPFD + TETKGTVLI TA+ELM+FS+GEE+ M+ +V+ IA TG VIVTGGKV ++A Sbjct: 242 YSCPFDGMITETKGTVLIKTAEELMNFSKGEENLMDAQVKAIAGTGANVIVTGGKVADIA 301 Query: 901 LHYANKYNLMVVKLSSKFDIRRVCKVTGATPIPKLSTPTQEELGYIASVRTEEIGKSTVV 1080 LHYANKYN+M+V+L+SK+D+RR+CK GAT +PKL+ P QEE+G+ SV E+G + VV Sbjct: 302 LHYANKYNIMLVRLNSKWDLRRLCKTVGATALPKLTPPVQEEMGHCDSVYLSEVGDTQVV 361 Query: 1081 LFDQDVKEGAISSIVVRGSMYDMMDEVERAIDDGVNTFKCLTKEPRYLAGAGATEIELAN 1260 +F + ++GAIS+IV+RGS ++MD++ERA+DDGVNTFK LT++ R + G GATEIELA Sbjct: 362 VFKHEKEDGAISTIVLRGSTDNLMDDIERAVDDGVNTFKVLTRDKRLVPGGGATEIELAK 421 Query: 1261 RLSDFANTQTGLVQYAIKQFVLAFESIVKVLCDNAGENATERIAELKAHHREGKTTFGII 1440 +++ + T GL QYAIK+F AFE+I + L +N+G A E I++L + H+EG G+ Sbjct: 422 QITSYGETCPGLEQYAIKKFAEAFEAIPRALAENSGVKANEVISKLYSVHQEGNKNVGLD 481 Query: 1441 STSEGRKVDDAVEHQILDTYTAKYWGIRFAADTAVTILRVDNIIM 1575 +E V D +E ILDTY KYW I+ A + AVT+LRVD IIM Sbjct: 482 IEAEVPAVKDMLEASILDTYLGKYWAIKLATNAAVTVLRVDQIIM 526
>sp|P78921|TCPQ_SCHPO Probable T-complex protein 1, theta subunit (TCP-1-theta) (CCT-theta) Length = 546 Score = 435 bits (1119), Expect = e-121 Identities = 225/527 (42%), Positives = 339/527 (64%), Gaps = 2/527 (0%) Frame = +1 Query: 1 AMHIPTAPGYLSMMKEGSRFYQGLEESVYRNIQACVALGQTTASTFGPQGMKKMVINHIG 180 A+ +P A G + +EG R QG+E++V RN A L + T ++ GP G K+V+NH+ Sbjct: 2 ALRVPKASGP-QLFREGYRIMQGVEDAVIRNCNAIRELSEITRTSLGPNGKNKIVVNHLQ 60 Query: 181 KKFVTKDASTILRELEVEHPAAKLMVMXXXXXXXXSGEATNFVIILCASLLEKAEYLLKM 360 + F+T DA+TI+RELEV HPAAKL+V G+A NFV++ LL KAE +++M Sbjct: 61 QTFLTNDAATIIRELEVIHPAAKLVVDATQQQENELGDAANFVVVFTGELLAKAENMIRM 120 Query: 361 GLSVPDVTEGFEIAAKHSLEILKDLVIGKVEDLREKEMIKKIVGTAVNSKIFGEESHISD 540 GL+ ++ +G+E+A H++E+L+++ K+E + ++ + K + T ++SK +G E +SD Sbjct: 121 GLTPLEIAKGYEMALSHTMEVLEEICADKIETVESEKELIKAIRTCISSKQYGNEDFLSD 180 Query: 541 LVVDACIRALPTS-SFFNVDNIRVVKILGSGTNASKVFNGMVFVREAQSVIKECKEAKVV 717 LV A + LP S FNVDNIRVVKI+GS S+V GMVF RE + + KEAKV Sbjct: 181 LVAKAILTVLPKDPSKFNVDNIRVVKIMGSSLYNSQVVKGMVFprePEGTVTRSKEAKVA 240 Query: 718 VFSCPFDQLQTETKGTVLINTAKELMDFSRGEEDQMEERVREIAETGCKVIVTGGKVGEL 897 VFSCP D QTETKGTVL++ A+E++DFS+GEE+ +E ++EI + G +V+VT G V +L Sbjct: 241 VFSCPLDISQTETKGTVLLHNAQEMLDFSKGEENLIESHIKEIYDAGVRVVVTSGNVNDL 300 Query: 898 ALHYANKYNLMVVKLSSKFDIRRVCKVTGATPIPKLSTPTQEELGYIASVRTEEIGKSTV 1077 LHY N++ ++V+++ SKF++RR+C+V GATP+ ++ P EE+G + V T EIG V Sbjct: 301 VLHYLNRFEILVIRVPSKFELRRLCRVVGATPLARMGVPMPEEMGSVDVVETIEIGGDRV 360 Query: 1078 VLFDQDVKEGAISSIVVRGSMYDMMDEVERAIDDGVNTFKCLTKEPRYLAGAGATEIELA 1257 +F Q ++IV+RG+ +D++ERAIDDGVN K L K+ R + GAGA++++L Sbjct: 361 TVFRQVEDITRTATIVLRGATKTYLDDLERAIDDGVNIVKALVKDNRLIFGAGASDMQLC 420 Query: 1258 NRLSDFANTQTGLVQYAIKQFVLAFESIVKVLCDNAGENATERIAEL-KAHHREGKTTFG 1434 RL G+ Q+AIKQ+ AFE + + + +NAG + T+ I++L AHH+E + G Sbjct: 421 IRLISVGEKTPGIYQHAIKQYGEAFEVVPRTISENAGLDPTDVISKLYAAHHKENGESIG 480 Query: 1435 IISTSEGRKVDDAVEHQILDTYTAKYWGIRFAADTAVTILRVDNIIM 1575 + E DA E I D AK IR A +T +T+L VD ++M Sbjct: 481 VDVECENDGTLDAKEAGIFDVLLAKKSAIRLATETVLTVLNVDQVVM 527
>sp|P47079|TCPQ_YEAST T-complex protein 1, theta subunit (TCP-1-theta) (CCT-theta) Length = 568 Score = 402 bits (1034), Expect = e-111 Identities = 222/548 (40%), Positives = 332/548 (60%), Gaps = 23/548 (4%) Frame = +1 Query: 1 AMHIPTAPGYLSMMKEGSRFYQGLEESVYRNIQACVALGQTTASTFGPQGMKKMVINHIG 180 ++ +P P + K+G Y + + ++I A L Q ++ GP G K+++NH+G Sbjct: 2 SLRLPQNPN-AGLFKQGYNSYSNADGQIIKSIAAIRELHQMCLTSMGPCGRNKIIVNHLG 60 Query: 181 KKFVTKDASTILRELEVEHPAAKLMVMXXXXXXXXSGEATNFVIILCASLLEKAEYLLKM 360 K +T DA+T+LREL++ HPA K++VM G+ TN V+IL LL +E L+ M Sbjct: 61 KIIITNDAATMLRELDIVHPAVKVLVMATEQQKIDMGDGTNLVMILAGELLNVSEKLISM 120 Query: 361 GLSVPDVTEGFEIAAKHSLEILKDLVIGKVEDLREKEMIKKIVGTAVNSKIFGEESHISD 540 GLS ++ +G+ +A K +L+ L ++V+G++ D +K + K++ ++SK +G E +S+ Sbjct: 121 GLSAVEIIQGYNMARKFTLKELDEMVVGEITDKNDKNELLKMIKPVISSKKYGSEDILSE 180 Query: 541 LVVDACIRALPTSS------FFNVDNIRVVKILGSGTNASKVFNGMVFVREAQSVIK--- 693 LV +A LP + +FNVD+IRVVKI+G + S V GMVF RE + +K Sbjct: 181 LVSEAVSHVLPVAQQAGEIPYFNVDSIRVVKIMGGSLSNSTVIKGMVFNREPEGHVKSLS 240 Query: 694 ECKEAKVVVFSCPFDQLQTETKGTVLINTAKELMDFSRGEEDQMEERVREIAETGCKVIV 873 E K+ KV VF+CP D TETKGTVL++ A+E++DFS+GEE Q++ ++EIA+ G + IV Sbjct: 241 EDKKHKVAVFTCPLDIANTETKGTVLLHNAQEMLDFSKGEEKQIDAMMKEIADMGVECIV 300 Query: 874 TGGKVGELALHYANKYNLMVVKLSSKFDIRRVCKVTGATPIPKLSTPTQEELGYIASVRT 1053 G VGELALHY N+Y ++V+K+ SKF++RR+C+V GATP+P+L PT EELG + +V+T Sbjct: 301 AGAGVGELALHYLNRYGILVLKVPSKFELRRLCRVCGATPLPRLGAPTPEELGLVETVKT 360 Query: 1054 EEIGKSTVVLFDQDVKE-GAISSIVVRGSMYDMMDEVERAIDDGVNTFKCLTKEP--RYL 1224 EIG V +F Q+ E S+I++RG+ + +D++ERAIDDGV K L K + L Sbjct: 361 MEIGGDRVTVFKQEQGEISRTSTIILRGATQNNLDDIERAIDDGVAAVKGLMKPSGGKLL 420 Query: 1225 AGAGATEIELANRLSDFANTQTGLVQYAIKQFVLAFESIVKVLCDNAGENATERIAELKA 1404 GAGATEIEL +R++ + GL+Q AIKQF +AFE + + L + AG + E + L A Sbjct: 421 PGAGATEIELISRITKYGERTPGLLQLAIKQFAVAFEVVPRTLAETAGLDVNEVLPNLYA 480 Query: 1405 HHREGKTTFGIISTSEGRK-----------VDDAVEHQILDTYTAKYWGIRFAADTAVTI 1551 H T G + T K V D E I D K + I A + A T+ Sbjct: 481 AH--NVTEPGAVKTDHLYKGVDIDGESDEGVKDIREENIYDMLATKKFAINVATEAATTV 538 Query: 1552 LRVDNIIM 1575 L +D IIM Sbjct: 539 LSIDQIIM 546
>sp|P47828|TCPQ_CANAL T-complex protein 1, theta subunit (TCP-1-theta) (CCT-theta) Length = 540 Score = 345 bits (884), Expect = 3e-94 Identities = 198/526 (37%), Positives = 301/526 (57%), Gaps = 1/526 (0%) Frame = +1 Query: 1 AMHIPTAPGYLSMMKEGSRFYQGLEESVYRNIQACVALGQTTASTFGPQGMKKMVINHIG 180 ++ +P AP + K+G + + ++ RN++A + ++ GP G K+++N +G Sbjct: 2 SLKLPQAPNS-GLFKQGYSSFSNADGAIIRNVEAVREIASILLTSMGPSGRNKIIVNKLG 60 Query: 181 KKFVTKDASTILRELEVEHPAAKLMVMXXXXXXXXSGEATNFVIILCASLLEKAEYLLKM 360 KKF+T DA+T+L ELE+ HP K+++ G+ TN VIIL L AE LL + Sbjct: 61 KKFITNDAATMLNELEIVHPVVKILIQASKQQEFEMGDNTNLVIILAGEFLNVAEKLLTL 120 Query: 361 GLSVPDVTEGFEIAAKHSLEILKDLVIGKVEDLREKEMIKKIVGTAVNSKIFGEESHISD 540 GL+V ++ +GF +A K ++ L +LV+ KVE E +++K V + +K +G E I+ Sbjct: 121 GLNVSEIIQGFNLANKFVMKTLDELVVEKVESF-ETDLLKA-VKPVIAAKQYGVEDTIAK 178 Query: 541 LVVDACIRALPTSSFFNVDNIRVVKILGSGTNASKVFNGMVFVREAQSVIKECKEAKVVV 720 LVVDA + SF NVDNIRVVK++G+ + S+V GMVF RE + +K + + Sbjct: 179 LVVDAVALVMKNGSF-NVDNIRVVKVMGASLSQSQVVKGMVFprePEGTVKNADQIQSCR 237 Query: 721 FSCPFDQLQTETKGTVLINTA-KELMDFSRGEEDQMEERVREIAETGCKVIVTGGKVGEL 897 P + +T ++ + R + + R+ G + ++ G VGEL Sbjct: 238 VYQPHRYFHHRNQRYSCSSTMPRKCLISPRAKNNSWTSCARKSTIQGLRWLLPGSSVGEL 297 Query: 898 ALHYANKYNLMVVKLSSKFDIRRVCKVTGATPIPKLSTPTQEELGYIASVRTEEIGKSTV 1077 ALHY NKY ++V+++ SKFD+RR+C+V GATP+P+L PT +E+G I + T+EIG V Sbjct: 298 ALHYLNKYGILVLRVPSKFDLRRICQVCGATPLPRLGAPTPDEMGEIDIIETKEIGGDRV 357 Query: 1078 VLFDQDVKEGAISSIVVRGSMYDMMDEVERAIDDGVNTFKCLTKEPRYLAGAGATEIELA 1257 +F QD ++IVVRG+ + +D++ERAIDDGVN+ K L K+ R L GAGA EIEL Sbjct: 358 TIFRQDESSSRTATIVVRGATQNNLDDIERAIDDGVNSIKGLIKDNRLLPGAGAVEIELM 417 Query: 1258 NRLSDFANTQTGLVQYAIKQFVLAFESIVKVLCDNAGENATERIAELKAHHREGKTTFGI 1437 R++ + +T GL+Q AIK F AFE I +VL + +G +++E +++L A H E Sbjct: 418 KRITAYQST-PGLLQLAIKSFAKAFEVIPRVLAETSGHDSSEILSKLHAAHAEDTGLRAG 476 Query: 1438 ISTSEGRKVDDAVEHQILDTYTAKYWGIRFAADTAVTILRVDNIIM 1575 I G D AV LD K I A D TIL +D IIM Sbjct: 477 IDIDSGEVADTAV----LDILATKKSAIDLAVDATNTILSIDQIIM 518
>sp|Q58405|THS_METJA Thermosome subunit (Chaperonin subunit) Length = 542 Score = 261 bits (667), Expect = 4e-69 Identities = 165/502 (32%), Positives = 272/502 (54%), Gaps = 8/502 (1%) Frame = +1 Query: 91 NIQACVALGQTTASTFGPQGMKKMVINHIGKKFVTKDASTILRELEVEHPAAKLMVMXXX 270 NI A + +T +T GP+GM KM+++ +G VT D TIL+E+ VEHPAAK+++ Sbjct: 27 NILAGRIIAETVRTTLGPKGMDKMLVDELGDIVVTNDGVTILKEMSVEHPAAKMLIEVAK 86 Query: 271 XXXXXSGEATNFVIILCASLLEKAEYLLKMGLSVPDVTEGFEIAAKHSLEILKDLVIGKV 450 G+ T +++ LL KAE LL + + G+E+A ++E LK I K Sbjct: 87 TQEKEVGDGTTTAVVIAGELLRKAEELLDQNIHPSVIINGYEMARNKAVEELKS--IAKE 144 Query: 451 EDLREKEMIKKIVGTAVNSKIFGEES---HISDLVVDACIRALPTSSFFNVDN--IRVVK 615 + EM+KKI T++ K G E ++++VV+A +RA+ VD I+V K Sbjct: 145 VKPEDTEMLKKIAMTSITGK--GAEKAREQLAEIVVEA-VRAVVDEETGKVDKDLIKVEK 201 Query: 616 ILGSGTNASKVFNGMVFVREAQS--VIKECKEAKVVVFSCPFDQLQTETKGTVLINTAKE 789 G+ +K+ G+V +E + + K+ + AK+ + +CP + +TET + I + Sbjct: 202 KEGAPIEETKLIRGVVIDKERVNPQMPKKVENAKIALLNCPIEVKETETDAEIRITDPAK 261 Query: 790 LMDFSRGEEDQMEERVREIAETGCKVIVTGGKVGELALHYANKYNLMVVKLSSKFDIRRV 969 LM+F EE +++ V +IA TG V+ + +LA HY K ++ V+ K D+ ++ Sbjct: 262 LMEFIEQEEKMIKDMVEKIAATGANVVFCQKGIDDLAQHYLAKKGILAVRRVKKSDMEKL 321 Query: 970 CKVTGATPIPKLSTPTQEELGYIASVRTEEIGKSTVVLFDQDVKEGAISSIVVRGSMYDM 1149 K TGA + K+ T E+LG V ++ ++ +Q A++ I+ RGS + Sbjct: 322 AKATGARIVTKIDDLTPEDLGEAGLVEERKVAGDAMIFVEQCKHPKAVT-ILARGSTEHV 380 Query: 1150 MDEVERAIDDGVNTFKCLTKEPRYLAGAGATEIELANRLSDFANTQTGLVQYAIKQFVLA 1329 ++EV RAIDD + KC +E + +AG GATEIELA RL FA + G Q A+K F A Sbjct: 381 VEEVARAIDDAIGVVKCALEEGKIVAGGGATEIELAKRLRKFAESVAGREQLAVKAFADA 440 Query: 1330 FESIVKVLCDNAGENATERIAELK-AHHREGKTTFGIISTSEGRKVDDAVEHQILDTYTA 1506 E I + L +N+G + + + +L+ AH +EG +G + EG +V D +E +++ Sbjct: 441 LEVIPRTLAENSGLDPIDMLVKLRAAHEKEGGEVYG-LDVFEG-EVVDMLEKGVVEPLKV 498 Query: 1507 KYWGIRFAADTAVTILRVDNII 1572 K I A + +V +LR+D++I Sbjct: 499 KTQAIDSATEASVMLLRIDDVI 520
>sp|O93624|THS_METTL Thermosome subunit (Chaperonin subunit) Length = 544 Score = 256 bits (654), Expect = 1e-67 Identities = 148/499 (29%), Positives = 270/499 (54%), Gaps = 5/499 (1%) Frame = +1 Query: 91 NIQACVALGQTTASTFGPQGMKKMVINHIGKKFVTKDASTILRELEVEHPAAKLMVMXXX 270 NI A +G+T ST GP+GM KM+++ +G VT D TIL+E+ VEHPAAK+++ Sbjct: 26 NILAGRIIGETVRSTLGPKGMDKMLVDDLGDIVVTNDGVTILKEMSVEHPAAKMLIEVAK 85 Query: 271 XXXXXSGEATNFVIILCASLLEKAEYLLKMGLSVPDVTEGFEIAAKHSLEILKDLVIGKV 450 G+ T +++ LL KAE LL + V +G+++A + + E+LK++ + Sbjct: 86 TQEKEVGDGTTTAVVIAGELLRKAEELLDQNVHPTIVIKGYQLAVQKAQEVLKEIAMDVK 145 Query: 451 EDLREKEMIKKIVGTAVNSKIFGEE---SHISDLVVDACIRALPTSSFFNVDNIRVVKIL 621 D +KE++ KI T++ K G E + +++V+A + S + D I++ K Sbjct: 146 AD--DKEILHKIAMTSITGK--GAEKAKEKLGEMIVEAVTAVVDESGKVDKDLIKIEKKE 201 Query: 622 GSGTNASKVFNGMVFVREAQS--VIKECKEAKVVVFSCPFDQLQTETKGTVLINTAKELM 795 G+ + +++ NG++ +E S + K+ + AK+ + +CP + +TET + I +LM Sbjct: 202 GASVDETELINGVLIDKERVSPQMPKKIENAKIALLNCPIEVKETETDAEIRITDPTKLM 261 Query: 796 DFSRGEEDQMEERVREIAETGCKVIVTGGKVGELALHYANKYNLMVVKLSSKFDIRRVCK 975 +F EE +++ V I +G V+ + +LA HY K ++ V+ K D+ ++ K Sbjct: 262 EFIEQEEKMLKDMVDTIKASGANVLFCQKGIDDLAQHYLAKEGILAVRRVKKSDMEKLSK 321 Query: 976 VTGATPIPKLSTPTQEELGYIASVRTEEIGKSTVVLFDQDVKEGAISSIVVRGSMYDMMD 1155 TGA + + E+LG V +I ++F ++ K ++++RG+ +++ Sbjct: 322 ATGANVVTNIKDLKAEDLGEAGIVEERKIA-GDAMIFVEECKHPKAVTMLIRGTTEHVIE 380 Query: 1156 EVERAIDDGVNTFKCLTKEPRYLAGAGATEIELANRLSDFANTQTGLVQYAIKQFVLAFE 1335 EV RA+DD + C ++ + +AG GA EIELA +L D+A +G Q A++ F A E Sbjct: 381 EVARAVDDAIGVVACTIEDGKIVAGGGAAEIELAMKLRDYAEGVSGREQLAVRAFADALE 440 Query: 1336 SIVKVLCDNAGENATERIAELKAHHREGKTTFGIISTSEGRKVDDAVEHQILDTYTAKYW 1515 + + L +NAG +A E + +L+A H EG + ++ G V++ E+ +++ K Sbjct: 441 VVPRTLAENAGLDAIEMLVKLRAKHAEGNNAYYGLNVFTG-DVENMTENGVVEPLRVKTQ 499 Query: 1516 GIRFAADTAVTILRVDNII 1572 I+ A + +LR+D++I Sbjct: 500 AIQSATEATEMLLRIDDVI 518
>sp|O26885|THSB_METTH Thermosome beta subunit (Thermosome subunit 2) (Chaperonin beta subunit) Length = 538 Score = 256 bits (653), Expect = 2e-67 Identities = 159/517 (30%), Positives = 280/517 (54%), Gaps = 5/517 (0%) Frame = +1 Query: 37 MMKEGSRFYQGLEESVYRNIQACVALGQTTASTFGPQGMKKMVINHIGKKFVTKDASTIL 216 ++ EG+ Y G ++ NI A L +T +T GP+GM KM+++ +G VT D TIL Sbjct: 10 VLPEGTSRYLG-RDAQRMNILAGKILAETVRTTLGPKGMDKMLVDSLGDIVVTNDGVTIL 68 Query: 217 RELEVEHPAAKLMVMXXXXXXXXSGEATNFVIILCASLLEKAEYLLKMGLSVPDVTEGFE 396 +E+++EHPAAK++V G+ T +I+ LL+KAE LL+M + + G+ Sbjct: 69 KEMDIEHPAAKMLVEVAKTQEDEVGDGTTTAVIIAGELLKKAENLLEMEIHPTIIAMGYR 128 Query: 397 IAAKHSLEILKDLVIGKVEDLREKEMIKKIVGTAVNSKIFGEE---SHISDLVVDACIRA 567 AA+ + EIL D+ I D +++ + K+ TA+ K G E +++L+VDA ++ Sbjct: 129 QAAEKAQEILDDIAI----DASDRDTLMKVAMTAMTGK--GTEKAREPLAELIVDA-VKQ 181 Query: 568 LPTSSFFNVDNIRVVKILGSGTNASKVFNGMVFVREA--QSVIKECKEAKVVVFSCPFDQ 741 + D+I++ K G+ + S + G++ +E + K+ + AK+ + +CP + Sbjct: 182 VEEDGEVEKDHIKIEKKEGAAVDDSTLVQGVIIDKERVHPGMPKKVENAKIALLNCPIEV 241 Query: 742 LQTETKGTVLINTAKELMDFSRGEEDQMEERVREIAETGCKVIVTGGKVGELALHYANKY 921 +TE + I ++ F EE + + V I +TG V+ + +LA HY K Sbjct: 242 KETEVDAEIRITDPSQMQAFIEQEEQMIRDMVNSIVDTGANVLFCQKGIDDLAQHYLAKA 301 Query: 922 NLMVVKLSSKFDIRRVCKVTGATPIPKLSTPTQEELGYIASVRTEEIGKSTVVLFDQDVK 1101 ++ V+ K D+ ++ K TGA + + + E+LG V ++I ++ F ++ K Sbjct: 302 GVLAVRRVKKSDMEKLSKATGANIVTNIEDLSPEDLGEAGVVSEKKISGEEMI-FVEECK 360 Query: 1102 EGAISSIVVRGSMYDMMDEVERAIDDGVNTFKCLTKEPRYLAGAGATEIELANRLSDFAN 1281 E +I+VRGS ++ EVERAI+D + ++ + +AG GA EIE+A RL D+A+ Sbjct: 361 EPKAVTILVRGSTEHVVSEVERAIEDAIGVVAATVEDGKVVAGGGAPEIEIAKRLKDYAD 420 Query: 1282 TQTGLVQYAIKQFVLAFESIVKVLCDNAGENATERIAELKAHHREGKTTFGIISTSEGRK 1461 + +G Q A+ F A E + K L +NAG ++ + + +L+A H E +T+ I +G K Sbjct: 421 SISGREQLAVSAFAEALEIVPKTLAENAGLDSIDVLVDLRAAHEE--STYMGIDVFDG-K 477 Query: 1462 VDDAVEHQILDTYTAKYWGIRFAADTAVTILRVDNII 1572 + D E +++ + K I+ AA+ A ILR+D++I Sbjct: 478 IVDMKEAGVIEPHRVKKQAIQSAAEAAEMILRIDDVI 514
>sp|O28045|THSA_ARCFU Thermosome alpha subunit (Thermosome subunit 1) (Chaperonin alpha subunit) Length = 545 Score = 255 bits (652), Expect = 2e-67 Identities = 165/518 (31%), Positives = 269/518 (51%), Gaps = 6/518 (1%) Frame = +1 Query: 37 MMKEGSRFYQGLEESVYRNIQACVALGQTTASTFGPQGMKKMVINHIGKKFVTKDASTIL 216 ++KEG++ G ++ NI A + + ST GP+GM KM+++ +G +T D TIL Sbjct: 11 ILKEGTQRTVG-RDAQRMNIMAARVIAEAVKSTLGPKGMDKMLVDSLGDVVITNDGVTIL 69 Query: 217 RELEVEHPAAKLMVMXXXXXXXXSGEATNFVIILCASLLEKAEYLLKMGLSVPDVTEGFE 396 +E++VEHPAAK+++ G+ T ++L LL+KAE LL + + G+ Sbjct: 70 KEMDVEHPAAKMIIEVAKTQDNEVGDGTTTAVVLAGELLKKAEELLDQDIHPTVIARGYR 129 Query: 397 IAAKHSLEILKDLVIGKVEDLREKEMIKKIVGTAVNSKIFGEE-SHISDLVVDACIR-AL 570 +AA ++EIL+ + + D+ ++E +KKI TA+ K H+S LVV+A R A Sbjct: 130 MAANKAVEILESIAMD--IDVEDEETLKKIAATAITGKHSEYALDHLSSLVVEAVKRVAE 187 Query: 571 PTSSFFNV--DNIRVVKILGSGTNASKVFNGMVFVREA--QSVIKECKEAKVVVFSCPFD 738 + V DNI++ K G +K+ NG+V +E + K K AK+ V + Sbjct: 188 KVDDRYKVDEDNIKLEKRQGGSVADTKLVNGIVIDKEVVHPGMPKRVKNAKIAVLKAALE 247 Query: 739 QLQTETKGTVLINTAKELMDFSRGEEDQMEERVREIAETGCKVIVTGGKVGELALHYANK 918 +TET + I +LM F EE ++E V +AE G V+ + +LA +Y K Sbjct: 248 VKETETDAEIRITDPDQLMKFIEQEEKMLKEMVDRLAEAGANVVFCQKGIDDLAQYYLAK 307 Query: 919 YNLMVVKLSSKFDIRRVCKVTGATPIPKLSTPTQEELGYIASVRTEEIGKSTVVLFDQDV 1098 ++ V+ + DI ++ K GA I L T +LG V ++G +V F + Sbjct: 308 AGILAVRRVKQSDIEKIAKACGAKIITDLREITSADLGEAELVEERKVGDEKMV-FIEGC 366 Query: 1099 KEGAISSIVVRGSMYDMMDEVERAIDDGVNTFKCLTKEPRYLAGAGATEIELANRLSDFA 1278 K +I++RG ++DEVER++ D + K + + +AG GA EIE+A ++ D+A Sbjct: 367 KNPKAVTILIRGGSEHVVDEVERSLQDAIKVVKTALESGKVVAGGGAPEIEVALKIRDWA 426 Query: 1279 NTQTGLVQYAIKQFVLAFESIVKVLCDNAGENATERIAELKAHHREGKTTFGIISTSEGR 1458 T G Q A + F A E I + L +N+G + + + EL+ H EGKTT+G+ S Sbjct: 427 PTLGGREQLAAEAFASALEVIPRALAENSGLDPIDILVELRKAHEEGKTTYGVDVFS--G 484 Query: 1459 KVDDAVEHQILDTYTAKYWGIRFAADTAVTILRVDNII 1572 +V E +L+ K I A + A+ ILR+D++I Sbjct: 485 EVACMKERGVLEPLKVKTQAITSATEVAIMILRIDDVI 522
>sp|P50016|THS_METKA Thermosome subunit (Chaperonin-like complex) (CLIC) Length = 545 Score = 249 bits (636), Expect = 2e-65 Identities = 153/518 (29%), Positives = 275/518 (53%), Gaps = 6/518 (1%) Frame = +1 Query: 37 MMKEGSRFYQGLEESVYRNIQACVALGQTTASTFGPQGMKKMVINHIGKKFVTKDASTIL 216 ++ EG + + G ++ NI A + +T +T GP GM KM+++ +G VT D TIL Sbjct: 13 ILPEGYQRFVG-RDAQRMNIMAARVVAETVRTTLGPMGMDKMLVDEMGDVVVTNDGVTIL 71 Query: 217 RELEVEHPAAKLMVMXXXXXXXXSGEATNFVIILCASLLEKAEYLLKMGLSVPDVTEGFE 396 E+++EHPAAK++V G+ T ++L LL KAE LL+ + + G+ Sbjct: 72 EEMDIEHPAAKMVVEVAKTQEDEVGDGTTTAVVLAGELLHKAEDLLQQDIHPTVIARGYR 131 Query: 397 IAAKHSLEILKDLVIGKVEDLREKEMIKKIVGTAVNSK-IFGEESHISDLVVDACIRALP 573 +A + + EIL++ I + D ++E +KKI TA+ K + ++++LVV A + Sbjct: 132 MAVEKAEEILEE--IAEEIDPDDEETLKKIAKTAMTGKGVEKARDYLAELVVKAVKQVAE 189 Query: 574 TSS---FFNVDNIRVVKILGSGTNASKVFNGMVFVREA--QSVIKECKEAKVVVFSCPFD 738 + D+I++ K G G +++ GMV +E + + + AK+ + +CP + Sbjct: 190 EEDGEIVIDTDHIKLEKKEGGGLEDTELVKGMVIDKERVHPGMPRRVENAKIALLNCPIE 249 Query: 739 QLQTETKGTVLINTAKELMDFSRGEEDQMEERVREIAETGCKVIVTGGKVGELALHYANK 918 +TET + I ++L F EE + E V +IAETG V+ + +LA HY K Sbjct: 250 VKETETDAEIRITDPEQLQAFIEEEERMLSEMVDKIAETGANVVFCQKGIDDLAQHYLAK 309 Query: 919 YNLMVVKLSSKFDIRRVCKVTGATPIPKLSTPTQEELGYIASVRTEEIGKSTVVLFDQDV 1098 ++ V+ K D++++ + TGA + + ++E+LG V +++ ++ F + Sbjct: 310 KGILAVRRVKKSDMQKLARATGARIVTNIDDLSEEDLGEAEVVEEKKVAGDKMI-FVEGC 368 Query: 1099 KEGAISSIVVRGSMYDMMDEVERAIDDGVNTFKCLTKEPRYLAGAGATEIELANRLSDFA 1278 K+ +I++RG ++DE ERAI+D + ++ + +AG GA E+E+A +L DFA Sbjct: 369 KDPKAVTILIRGGTEHVVDEAERAIEDAIGVVAAALEDGKVVAGGGAPEVEVARQLRDFA 428 Query: 1279 NTQTGLVQYAIKQFVLAFESIVKVLCDNAGENATERIAELKAHHREGKTTFGIISTSEGR 1458 + G Q A++ F A E I + L +N+G + + + +L+A H +G+ T G I +G Sbjct: 429 DGVEGREQLAVEAFADALEIIPRTLAENSGLDPIDVLVQLRAKHEDGQVTAG-IDVYDG- 486 Query: 1459 KVDDAVEHQILDTYTAKYWGIRFAADTAVTILRVDNII 1572 V D +E +++ K + A + A ILR+D++I Sbjct: 487 DVKDMLEEGVVEPLRVKTQALASATEAAEMILRIDDVI 524
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 182,631,777
Number of Sequences: 369166
Number of extensions: 3592118
Number of successful extensions: 10997
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9930
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10738
length of database: 68,354,980
effective HSP length: 116
effective length of database: 46,925,720
effective search space used: 21257351160
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail