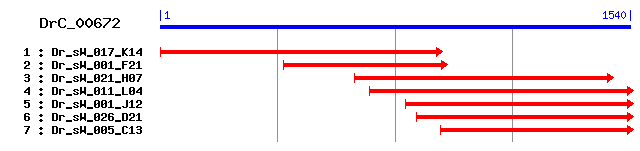
DrC_00672
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00672 (1533 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P38657|PDIA3_BOVIN Protein disulfide-isomerase A3 precur... 454 e-127 sp|P30101|PDIA3_HUMAN Protein disulfide-isomerase A3 precur... 454 e-127 sp|P27773|PDIA3_MOUSE Protein disulfide-isomerase A3 precur... 446 e-124 sp|P11598|PDIA3_RAT Protein disulfide-isomerase A3 precurso... 440 e-123 sp|P38658|ERP60_SCHMA Probable protein disulfide-isomerase ... 375 e-103 sp|P13667|PDIA4_HUMAN Protein disulfide-isomerase A4 precur... 350 6e-96 sp|P08003|PDIA4_MOUSE Protein disulfide-isomerase A4 precur... 346 1e-94 sp|P38659|PDIA4_RAT Protein disulfide-isomerase A4 precurso... 340 7e-93 sp|P34329|PDIA4_CAEEL Probable protein disulfide-isomerase ... 324 4e-88 sp|P55059|PDI_HUMIN Protein disulfide-isomerase precursor (... 274 6e-73
>sp|P38657|PDIA3_BOVIN Protein disulfide-isomerase A3 precursor (Disulfide isomerase ER-60) (ERP60) (58 kDa microsomal protein) (P58) (ERp57) Length = 505 Score = 454 bits (1169), Expect = e-127 Identities = 228/458 (49%), Positives = 307/458 (67%), Gaps = 7/458 (1%) Frame = +3 Query: 6 NFEQSIKE---HSVILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETK 176 NFE I + ++LVEF+APWCGHCKKLAPEYE AAT LK +P+AKVDCTA T Sbjct: 34 NFESRITDTGSSGLMLVEFFAPWCGHCKKLAPEYEAAATRLKGI---VPLAKVDCTANTN 90 Query: 177 TCEKHGVSGYPTLKIFKNGQFAFDYSGERSSDGIIKYMISKSGPSSKEVKTEENFKKFIA 356 TC K+GVSGYPTLKIF++G+ + Y G R++DGI+ ++ ++GP+S +K+EE F+KFI+ Sbjct: 91 TCNKYGVSGYPTLKIFRDGEESGAYDGPRTADGIVSHLKKQAGPASVPLKSEEEFEKFIS 150 Query: 357 STDPVVVGFFASESSDLYKAFLSVAEKLNEDFRVAHVFDESFESKIKFRKTTFVIYHPQK 536 D VVGFF S+ + FL A L +++R AH ES +K ++ P Sbjct: 151 DKDASVVGFFKDLFSEAHSEFLKAASNLRDNYRFAHTNVESLVNKYDDDGEGITLFRPSH 210 Query: 537 MANKFEESQIKYKGDASEISN*NMG*RKNNGVWSV*EQVPT*N--ISHPNLLLVALYSVD 710 + NKFE+ + Y ++N ++ + + N + LL+A Y VD Sbjct: 211 LTNKFEDKTVAYTEQKMTSGKIKRFIQEN--IFGICPHMTEDNKDLLQGKDLLIAYYDVD 268 Query: 711 YEKNPKGTNYWRNRIMKVAKKFVDSGKSVHFAISNSQEQSHELSEFGIESPDSNKVYIVA 890 YEKN KG+NYWRNR+M VAKKF+D+G+ +HFA+++ + SHELS+FG+ES + Sbjct: 269 YEKNAKGSNYWRNRVMMVAKKFLDAGQKLHFAVASRKTFSHELSDFGLESTTGEIPVVAV 328 Query: 891 RDEKNQKFKMAEDFTME--AFEKFINDFLNEQVPVYLKSETIPEKNDEPVKVVVAKNFDA 1064 R K +KF M E+F+ + A E+F+ D+ + + YLKSE IPE ND PVKVVVA+NFD Sbjct: 329 RTAKGEKFVMQEEFSRDGKALERFLEDYFDGNLKRYLKSEPIPESNDGPVKVVVAENFDE 388 Query: 1065 LVNDPTKDVLIEFYAPWCGHCKSLAPKYEELAKKLSGESDIVIAKMDATANDVPSPYEVT 1244 +VN+ KDVLIEFYAPWCGHCK+L PKY+EL +KL + +IVIAKMDATANDVPSPYEV Sbjct: 389 IVNNENKDVLIEFYAPWCGHCKNLEPKYKELGEKLRKDPNIVIAKMDATANDVPSPYEVR 448 Query: 1245 GFPTIYFAPKNSKNSPKSYNGGREVDDMLKYIASESTS 1358 GFPTIYF+P N K +PK Y GGRE+ D + Y+ E+T+ Sbjct: 449 GFPTIYFSPANKKQNPKKYEGGRELSDFISYLKREATN 486
>sp|P30101|PDIA3_HUMAN Protein disulfide-isomerase A3 precursor (Disulfide isomerase ER-60) (ERp60) (58 kDa microsomal protein) (p58) (ERp57) (58 kDa glucose regulated protein) Length = 505 Score = 454 bits (1167), Expect = e-127 Identities = 227/458 (49%), Positives = 308/458 (67%), Gaps = 7/458 (1%) Frame = +3 Query: 6 NFEQSIKEHS---VILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETK 176 NFE I + ++LVEF+APWCGHCK+LAPEYE AAT LK +P+AKVDCTA T Sbjct: 34 NFESRISDTGSAGLMLVEFFAPWCGHCKRLAPEYEAAATRLKGI---VPLAKVDCTANTN 90 Query: 177 TCEKHGVSGYPTLKIFKNGQFAFDYSGERSSDGIIKYMISKSGPSSKEVKTEENFKKFIA 356 TC K+GVSGYPTLKIF++G+ A Y G R++DGI+ ++ ++GP+S ++TEE FKKFI+ Sbjct: 91 TCNKYGVSGYPTLKIFRDGEEAGAYDGPRTADGIVSHLKKQAGPASVPLRTEEEFKKFIS 150 Query: 357 STDPVVVGFFASESSDLYKAFLSVAEKLNEDFRVAHVFDESFESKIKFRKTTFVIYHPQK 536 D +VGFF S+ + FL A L +++R AH ES ++ +++ P Sbjct: 151 DKDASIVGFFDDSFSEAHSEFLKAASNLRDNYRFAHTNVESLVNEYDDNGEGIILFRPSH 210 Query: 537 MANKFEESQIKYKGDASEISN*NMG*RKNNGVWSV*EQVPT*N--ISHPNLLLVALYSVD 710 + NKFE+ + Y ++N ++ + + N + LL+A Y VD Sbjct: 211 LTNKFEDKTVAYTEQKMTSGKIKKFIQEN--IFGICPHMTEDNKDLIQGKDLLIAYYDVD 268 Query: 711 YEKNPKGTNYWRNRIMKVAKKFVDSGKSVHFAISNSQEQSHELSEFGIESPDSNKVYIVA 890 YEKN KG+NYWRNR+M VAKKF+D+G ++FA+++ + SHELS+FG+ES + Sbjct: 269 YEKNAKGSNYWRNRVMMVAKKFLDAGHKLNFAVASRKTFSHELSDFGLESTAGEIPVVAI 328 Query: 891 RDEKNQKFKMAEDFTME--AFEKFINDFLNEQVPVYLKSETIPEKNDEPVKVVVAKNFDA 1064 R K +KF M E+F+ + A E+F+ D+ + + YLKSE IPE ND PVKVVVA+NFD Sbjct: 329 RTAKGEKFVMQEEFSRDGKALERFLQDYFDGNLKRYLKSEPIPESNDGPVKVVVAENFDE 388 Query: 1065 LVNDPTKDVLIEFYAPWCGHCKSLAPKYEELAKKLSGESDIVIAKMDATANDVPSPYEVT 1244 +VN+ KDVLIEFYAPWCGHCK+L PKY+EL +KLS + +IVIAKMDATANDVPSPYEV Sbjct: 389 IVNNENKDVLIEFYAPWCGHCKNLEPKYKELGEKLSKDPNIVIAKMDATANDVPSPYEVR 448 Query: 1245 GFPTIYFAPKNSKNSPKSYNGGREVDDMLKYIASESTS 1358 GFPTIYF+P N K +PK Y GGRE+ D + Y+ E+T+ Sbjct: 449 GFPTIYFSPANKKLNPKKYEGGRELSDFISYLQREATN 486
>sp|P27773|PDIA3_MOUSE Protein disulfide-isomerase A3 precursor (Disulfide isomerase ER-60) (ERp60) (58 kDa microsomal protein) (p58) (ERp57) Length = 504 Score = 446 bits (1146), Expect = e-124 Identities = 226/458 (49%), Positives = 308/458 (67%), Gaps = 7/458 (1%) Frame = +3 Query: 6 NFEQSIKEHS---VILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETK 176 NFE + + ++LVEF+APWCGHCK+LAPEYE AAT LK +P+AKVDCTA T Sbjct: 34 NFESRVSDTGSAGLMLVEFFAPWCGHCKRLAPEYEAAATRLKI----VPLAKVDCTANTN 89 Query: 177 TCEKHGVSGYPTLKIFKNGQFAFDYSGERSSDGIIKYMISKSGPSSKEVKTEENFKKFIA 356 TC K+GVSGYPTLKIF+ G+ A Y G R++DGI+ ++ ++GP+S ++TEE FKKFI+ Sbjct: 90 TCNKYGVSGYPTLKIFRAGEEAGAYDGPRTADGIVSHLKKQAGPASVPLRTEEEFKKFIS 149 Query: 357 STDPVVVGFFASESSDLYKAFLSVAEKLNEDFRVAHVFDESFESKIKFRKTTFVIYHPQK 536 D VVGFF SD + FL A L +++R AH ES + I+ P Sbjct: 150 DKDASVVGFFRDLFSDGHSEFLKAASNLRDNYRFAHTNIESLVKEYDDNGEGITIFRPLH 209 Query: 537 MANKFEESQIKYKGDASEISN*NMG*RKNNGVWSV*EQVPT*N--ISHPNLLLVALYSVD 710 +ANKFE+ + Y ++++ + + ++ + + N + LL A Y VD Sbjct: 210 LANKFEDKTVAYT--EKKMTSAKIKKFIQDSIFGLCPHMTEDNKDLIQGKDLLTAYYDVD 267 Query: 711 YEKNPKGTNYWRNRIMKVAKKFVDSGKSVHFAISNSQEQSHELSEFGIESPDSNKVYIVA 890 YEKN KG+NYWRNR+M VAKKF+D+G ++FA+++ + SHELS+F +ES + Sbjct: 268 YEKNAKGSNYWRNRVMMVAKKFLDAGHKLNFAVASRKTFSHELSDFSLESTTGEVPVVAI 327 Query: 891 RDEKNQKFKMAEDFTME--AFEKFINDFLNEQVPVYLKSETIPEKNDEPVKVVVAKNFDA 1064 R K +KF M E+F+ + A E+F+ ++ + + YLKSE IPE N+ PVKVVVA+NFD Sbjct: 328 RTAKGEKFVMQEEFSRDGKALEQFLQEYFDGNLKRYLKSEPIPESNEGPVKVVVAENFDD 387 Query: 1065 LVNDPTKDVLIEFYAPWCGHCKSLAPKYEELAKKLSGESDIVIAKMDATANDVPSPYEVT 1244 +VN+ KDVLIEFYAPWCGHCK+L PKY+EL +KLS + +IVIAKMDATANDVPSPYEV Sbjct: 388 IVNEEDKDVLIEFYAPWCGHCKNLEPKYKELGEKLSKDPNIVIAKMDATANDVPSPYEVK 447 Query: 1245 GFPTIYFAPKNSKNSPKSYNGGREVDDMLKYIASESTS 1358 GFPTIYF+P N K +PK Y GGRE++D + Y+ E+T+ Sbjct: 448 GFPTIYFSPANKKLTPKKYEGGRELNDFISYLQREATN 485
Score = 68.9 bits (167), Expect = 4e-11
Identities = 37/104 (35%), Positives = 63/104 (60%), Gaps = 3/104 (2%)
Frame = +3
Query: 1050 KNFDALVNDPTKD--VLIEFYAPWCGHCKSLAPKYEELAKKLSGESDIVIAKMDATAN-D 1220
+NF++ V+D +L+EF+APWCGHCK LAP+YE A +L + +AK+D TAN +
Sbjct: 33 ENFESRVSDTGSAGLMLVEFFAPWCGHCKRLAPEYEAAATRL---KIVPLAKVDCTANTN 89
Query: 1221 VPSPYEVTGFPTIYFAPKNSKNSPKSYNGGREVDDMLKYIASES 1352
+ Y V+G+PT+ + +Y+G R D ++ ++ ++
Sbjct: 90 TCNKYGVSGYPTLKIF--RAGEEAGAYDGPRTADGIVSHLKKQA 131
Score = 64.3 bits (155), Expect = 9e-10
Identities = 42/123 (34%), Positives = 61/123 (49%), Gaps = 3/123 (2%)
Frame = +3
Query: 6 NFEQSIKEHSV-ILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETKTC 182
NF+ + E +L+EFYAPWCGHCK L P+Y+ L + DP I IAK+D TA
Sbjct: 384 NFDDIVNEEDKDVLIEFYAPWCGHCKNLEPKYKELGEKLSK-DPNIVIAKMDATA-NDVP 441
Query: 183 EKHGVSGYPTLKIFKNGQ--FAFDYSGERSSDGIIKYMISKSGPSSKEVKTEENFKKFIA 356
+ V G+PT+ + Y G R + I Y + + + ++ E+ KK A
Sbjct: 442 SPYEVKGFPTIYFSPANKKLTPKKYEGGRELNDFISY-LQREATNPPIIQEEKPKKKKKA 500
Query: 357 STD 365
D
Sbjct: 501 QED 503
>sp|P11598|PDIA3_RAT Protein disulfide-isomerase A3 precursor (Disulfide isomerase ER-60) (ERp60) (58 kDa microsomal protein) (p58) (ERp57) (HIP-70) (Q-2) Length = 505 Score = 440 bits (1132), Expect = e-123 Identities = 224/458 (48%), Positives = 307/458 (67%), Gaps = 7/458 (1%) Frame = +3 Query: 6 NFEQSIKEHS---VILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETK 176 NFE + + ++LVEF+APWCGHCK+LAPEYE AAT LK +P+AKVDCTA T Sbjct: 34 NFESRVSDTGSAGLMLVEFFAPWCGHCKRLAPEYEAAATRLKGI---VPLAKVDCTANTN 90 Query: 177 TCEKHGVSGYPTLKIFKNGQFAFDYSGERSSDGIIKYMISKSGPSSKEVKTEENFKKFIA 356 TC K+GVSGYPTLKIF++G+ A Y G R++DGI+ ++ ++GP+S ++TE+ FKKFI+ Sbjct: 91 TCNKYGVSGYPTLKIFRDGEEAGAYDGPRTADGIVSHLKKQAGPASVPLRTEDEFKKFIS 150 Query: 357 STDPVVVGFFASESSDLYKAFLSVAEKLNEDFRVAHVFDESFESKIKFRKTTFVIYHPQK 536 D VVGFF SD + FL A L +++R AH ES + I+ P Sbjct: 151 DKDASVVGFFRDLFSDGHSEFLKAASNLRDNYRFAHTNVESLVKEYDDNGEGITIFRPLH 210 Query: 537 MANKFEESQIKYKGDASEISN*NMG*RKNNGVWSV*EQVPT*N--ISHPNLLLVALYSVD 710 +ANKFE+ + Y ++++ + ++ + + N + LL A Y VD Sbjct: 211 LANKFEDKIVAYT--EKKMTSGKIKKFIQESIFGLCPHMTEDNKDLIQGKDLLTAYYDVD 268 Query: 711 YEKNPKGTNYWRNRIMKVAKKFVDSGKSVHFAISNSQEQSHELSEFGIESPDSNKVYIVA 890 YEKN KG+NYWRNR+M VAK F+D+G ++FA+++ + SHELS+FG+ES + Sbjct: 269 YEKNTKGSNYWRNRVMMVAKTFLDAGHKLNFAVASRKTFSHELSDFGLESTTGEIPVVAI 328 Query: 891 RDEKNQKFKMAEDFTME--AFEKFINDFLNEQVPVYLKSETIPEKNDEPVKVVVAKNFDA 1064 R K +KF M E+F+ + A E+F+ ++ + + YLKSE IPE N+ PVKVVVA++FD Sbjct: 329 RTAKGEKFVMQEEFSRDGKALERFLQEYFDGNLKRYLKSEPIPETNEGPVKVVVAESFDD 388 Query: 1065 LVNDPTKDVLIEFYAPWCGHCKSLAPKYEELAKKLSGESDIVIAKMDATANDVPSPYEVT 1244 +VN KDVLIEFYAPWCGHCK+L PKY+EL +KLS + +IVIAKMDATANDVPSPYEV Sbjct: 389 IVNAEDKDVLIEFYAPWCGHCKNLEPKYKELGEKLSKDPNIVIAKMDATANDVPSPYEVK 448 Query: 1245 GFPTIYFAPKNSKNSPKSYNGGREVDDMLKYIASESTS 1358 GFPTIYF+P N K +PK Y GGRE++D + Y+ E+T+ Sbjct: 449 GFPTIYFSPANKKLTPKKYEGGRELNDFISYLQREATN 486
Score = 72.0 bits (175), Expect = 4e-12
Identities = 43/127 (33%), Positives = 73/127 (57%), Gaps = 3/127 (2%)
Frame = +3
Query: 981 VPVYLKSETIPEKNDEPVKVVVAKNFDALVNDPTKD--VLIEFYAPWCGHCKSLAPKYEE 1154
V + L S + +D V + +NF++ V+D +L+EF+APWCGHCK LAP+YE
Sbjct: 12 VALLLASALLASASD--VLELTDENFESRVSDTGSAGLMLVEFFAPWCGHCKRLAPEYEA 69
Query: 1155 LAKKLSGESDIVIAKMDATAN-DVPSPYEVTGFPTIYFAPKNSKNSPKSYNGGREVDDML 1331
A +L G + +AK+D TAN + + Y V+G+PT+ + +Y+G R D ++
Sbjct: 70 AATRLKG--IVPLAKVDCTANTNTCNKYGVSGYPTLKIFRDGEEAG--AYDGPRTADGIV 125
Query: 1332 KYIASES 1352
++ ++
Sbjct: 126 SHLKKQA 132
Score = 62.4 bits (150), Expect = 3e-09
Identities = 40/115 (34%), Positives = 57/115 (49%), Gaps = 2/115 (1%)
Frame = +3
Query: 27 EHSVILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETKTCEKHGVSGY 206
E +L+EFYAPWCGHCK L P+Y+ L + DP I IAK+D TA + V G+
Sbjct: 393 EDKDVLIEFYAPWCGHCKNLEPKYKELGEKLSK-DPNIVIAKMDATA-NDVPSPYEVKGF 450
Query: 207 PTLKIFKNGQ--FAFDYSGERSSDGIIKYMISKSGPSSKEVKTEENFKKFIASTD 365
PT+ + Y G R + I Y + + + ++ E+ KK A D
Sbjct: 451 PTIYFSPANKKLTPKKYEGGRELNDFISY-LQREATNPPIIQEEKPKKKKKAQED 504
>sp|P38658|ERP60_SCHMA Probable protein disulfide-isomerase ER-60 precursor (ERP60) Length = 484 Score = 375 bits (963), Expect = e-103 Identities = 197/468 (42%), Positives = 286/468 (61%), Gaps = 1/468 (0%) Frame = +3 Query: 6 NFEQSIKEHSVILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETKTCE 185 NF +K V LV+FYAPWCGHCKKLAPE+ AA + + + + KVDCT + C Sbjct: 26 NFHSELKSIPVALVKFYAPWCGHCKKLAPEFTSAAQIISGKTNDVKLVKVDCTTQESICS 85 Query: 186 KHGVSGYPTLKIFKNGQFAFDYSGERSSDGIIKYMISKSGPSSKEVKTEENFKKFIASTD 365 + GVSGYPTLKIF+NG +Y+G R+++GI YMIS++GP SKEV T + + ++ Sbjct: 86 EFGVSGYPTLKIFRNGDLDGEYNGPRNANGIANYMISRAGPVSKEVSTVSDVENVLSDDK 145 Query: 366 PVVVGFFASESSDLYKAFLSVAEKLNEDFRVAHVFDESFESKIKFRKTTFVIYHPQKMAN 545 P V F S S L K F+++A+ + +D H + F + +Y P+++ Sbjct: 146 PTVFAFVKSSSDPLIKTFMALAKSMVDDAVFCHSHNNLFVTP---SDNELRVYLPKRLRT 202 Query: 546 KFEESQIKYKGDASEISN*NMG*RKNNGVWSV*EQVPT*NISHPNLLLVALYS-VDYEKN 722 KFE+ YKG+ SN + +G V + P+ N LV LY+ + Sbjct: 203 KFEDDFAVYKGELE--SNNIKDWIRKHGQGLVGYRSPSNTFYFENSDLVVLYNNQSIDSY 260 Query: 723 PKGTNYWRNRIMKVAKKFVDSGKSVHFAISNSQEQSHELSEFGIESPDSNKVYIVARDEK 902 P G Y RNR++K K K++ FA S + + S+E+S++GIE+ +K+ V K Sbjct: 261 PSGVKYLRNRVLKTLKDNPQKFKNLVFAYSFADDFSYEISDYGIEA---DKLPAVVIQSK 317 Query: 903 NQKFKMAEDFTMEAFEKFINDFLNEQVPVYLKSETIPEKNDEPVKVVVAKNFDALVNDPT 1082 ++K+K+ E F+++AF F+N F + + ++KSE +P + VK +VA NFD +VN+ Sbjct: 318 DKKYKL-EKFSLDAFSDFLNKFEDGLLTPHVKSEPLPTDDSSAVKKLVALNFDEIVNNEE 376 Query: 1083 KDVLIEFYAPWCGHCKSLAPKYEELAKKLSGESDIVIAKMDATANDVPSPYEVTGFPTIY 1262 KDV++ F+A WCGHCK+L PKYEE A K+ E ++V+A MDATANDVPSPY+V GFPTIY Sbjct: 377 KDVMVVFHAGWCGHCKNLMPKYEEAASKVKNEPNLVLAAMDATANDVPSPYQVRGFPTIY 436 Query: 1263 FAPKNSKNSPKSYNGGREVDDMLKYIASESTSSLKSYDRQGRVRKSDL 1406 F PK K+SP SY GGR+ +D++KY+A E+T L YDR G +KS+L Sbjct: 437 FVPKGKKSSPVSYEGGRDTNDIIKYLAREATEELIGYDRSGNPKKSEL 484
>sp|P13667|PDIA4_HUMAN Protein disulfide-isomerase A4 precursor (Protein ERp-72) (ERp72) Length = 645 Score = 350 bits (898), Expect = 6e-96 Identities = 192/466 (41%), Positives = 276/466 (59%), Gaps = 16/466 (3%) Frame = +3 Query: 6 NFEQSIKEHSVILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETKTCE 185 NF++ + + +ILVEFYAPWCGHCKKLAPEYE AA L ++ PPIP+AKVD TAET + Sbjct: 186 NFDEVVNDADIILVEFYAPWCGHCKKLAPEYEKAAKELSKRSPPIPLAKVDATAETDLAK 245 Query: 186 KHGVSGYPTLKIFKNGQFAFDYSGERSSDGIIKYMISKSGPSSKEVKTEENFKKFIASTD 365 + VSGYPTLKIF+ G+ +DY+G R GI+ YMI +SGP SKE+ T + ++F+ D Sbjct: 246 RFDVSGYPTLKIFRKGR-PYDYNGpreKYGIVDYMIEQSGPPSKEILTLKQVQEFLKDGD 304 Query: 366 PVVV-GFFASESSDLYKAFLSVAEKLNEDFRVAHVFDESFESKIKFRKTTFVIYHPQKMA 542 V++ G F ES Y+ + A L ED++ H F +K + V+ P+K Sbjct: 305 DVIIIGVFKGESDPAYQQYQDAANNLREDYKFHHTFSTEIAKFLKVSQGQLVVMQPEKFQ 364 Query: 543 NKFE--ESQIKYKGDASEISN*N---------MG*RKNNGVWSV*EQVPT*NISHPNLLL 689 +K+E + +G + + + +G RK + + P L+ Sbjct: 365 SKYEPRSHMMDVQGSTQDSAIKDFVLKYALPLVGHRKVSNDAKRYTRRP---------LV 415 Query: 690 VALYSVDYEKNPKG-TNYWRNRIMKVAKKFVDSGKSVHFAISNSQEQSHELSEFGIESPD 866 V YSVD+ + + T +WR+++++VAK F + FAI++ ++ + E+ + G+ S Sbjct: 416 VVYYSVDFSFDYRAATQFWRSKVLEVAKDFPE----YTFAIADEEDYAGEVKDLGL-SES 470 Query: 867 SNKVYIVARDEKNQKFKMA-EDFTMEAFEKFINDFLNEQVPVYLKSETIPEKNDEPVKVV 1043 V DE +KF M E+F + +F+ F ++ +KS+ +P+ N PVKVV Sbjct: 471 GEDVNAAILDESGKKFAMEPEEFDSDTLREFVTAFKKGKLKPVIKSQPVPKNNKGPVKVV 530 Query: 1044 VAKNFDALVNDPTKDVLIEFYAPWCGHCKSLAPKYEELAKKLSGESDIVIAKMDATANDV 1223 V K FD++V DP KDVLIEFYAPWCGHCK L P Y LAKK G+ +VIAKMDATANDV Sbjct: 531 VGKTFDSIVMDPKKDVLIEFYAPWCGHCKQLEPVYNSLAKKYKGQKGLVIAKMDATANDV 590 Query: 1224 PSP-YEVTGFPTIYFAPKNSKNSPKSYNGG-REVDDMLKYIASEST 1355 PS Y+V GFPTIYFAP K +P + GG R+++ + K+I +T Sbjct: 591 PSDRYKVEGFPTIYFAPSGDKKNPVKFEGGDRDLEHLSKFIEEHAT 636
Score = 120 bits (302), Expect = 8e-27
Identities = 59/135 (43%), Positives = 83/135 (61%), Gaps = 6/135 (4%)
Frame = +3
Query: 3 SNFEQSIKEHSVILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETKTC 182
+NF+ + + +L+EFYAPWCGHCK+ APEYE A LK +DPPIP+AK+D T+ +
Sbjct: 70 ANFDNFVADKDTVLLEFYAPWCGHCKQFAPEYEKIANILKDKDPPIPVAKIDATSASVLA 129
Query: 183 EKHGVSGYPTLKIFKNGQFAFDYSGERSSDGIIKYMISKSGPSSKE------VKTEENFK 344
+ VSGYPT+KI K GQ A DY G R+ + I+ + S P V T+ENF
Sbjct: 130 SRFDVSGYPTIKILKKGQ-AVDYEGSRTQEEIVAKVREVSQPDWTPPPEVTLVLTKENFD 188
Query: 345 KFIASTDPVVVGFFA 389
+ + D ++V F+A
Sbjct: 189 EVVNDADIILVEFYA 203
Score = 92.8 bits (229), Expect = 2e-18
Identities = 51/111 (45%), Positives = 71/111 (63%), Gaps = 2/111 (1%)
Frame = +3
Query: 1026 EPVKVVVAKNFDALVNDPTKDVLIEFYAPWCGHCKSLAPKYEELAKKLSGES-DIVIAKM 1202
E V+ +NFD +VND +L+EFYAPWCGHCK LAP+YE+ AK+LS S I +AK+
Sbjct: 177 EVTLVLTKENFDEVVND-ADIILVEFYAPWCGHCKKLAPEYEKAAKELSKRSPPIPLAKV 235
Query: 1203 DATA-NDVPSPYEVTGFPTIYFAPKNSKNSPKSYNGGREVDDMLKYIASES 1352
DATA D+ ++V+G+PT+ K P YNG RE ++ Y+ +S
Sbjct: 236 DATAETDLAKRFDVSGYPTLKIF---RKGRPYDYNGpreKYGIVDYMIEQS 283
Score = 76.6 bits (187), Expect = 2e-13
Identities = 47/122 (38%), Positives = 70/122 (57%), Gaps = 3/122 (2%)
Frame = +3
Query: 996 KSETIPEKNDEPVKVVVAKNFDALVNDPTKD-VLIEFYAPWCGHCKSLAPKYEELAKKLS 1172
+ + + K + V V+ NFD V D KD VL+EFYAPWCGHCK AP+YE++A L
Sbjct: 52 EEDDLEVKEENGVLVLNDANFDNFVAD--KDTVLLEFYAPWCGHCKQFAPEYEKIANILK 109
Query: 1173 G-ESDIVIAKMDAT-ANDVPSPYEVTGFPTIYFAPKNSKNSPKSYNGGREVDDMLKYIAS 1346
+ I +AK+DAT A+ + S ++V+G+PTI K Y G R ++++ +
Sbjct: 110 DKDPPIPVAKIDATSASVLASRFDVSGYPTIKIL---KKGQAVDYEGSRTQEEIVAKVRE 166
Query: 1347 ES 1352
S
Sbjct: 167 VS 168
>sp|P08003|PDIA4_MOUSE Protein disulfide-isomerase A4 precursor (Protein ERp-72) (ERp72) Length = 638 Score = 346 bits (887), Expect = 1e-94 Identities = 192/466 (41%), Positives = 271/466 (58%), Gaps = 16/466 (3%) Frame = +3 Query: 6 NFEQSIKEHSVILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETKTCE 185 NF+ + +ILVEFYAPWCGHCKKLAPEYE AA L ++ PPIP+AKVD T +T + Sbjct: 179 NFDDVVNNADIILVEFYAPWCGHCKKLAPEYEKAAKELSKRSPPIPLAKVDATEQTDLAK 238 Query: 186 KHGVSGYPTLKIFKNGQFAFDYSGERSSDGIIKYMISKSGPSSKEVKTEENFKKFIASTD 365 + VSGYPTLKIF+ G+ FDY+G R GI+ YMI +SGP SKE+ T + ++F+ D Sbjct: 239 RFDVSGYPTLKIFRKGR-PFDYNGpreKYGIVDYMIEQSGPPSKEILTLKQVQEFLKDGD 297 Query: 366 PVVV-GFFASESSDLYKAFLSVAEKLNEDFRVAHVFDESFESKIKFRKTTFVIYHPQKMA 542 VV+ G F + Y + A L ED++ H F +K V+ HP+K Sbjct: 298 DVVIIGLFQGDGDPAYLQYQDAANNLREDYKFHHTFSPEIAKFLKVSLGKLVLTHPEKFQ 357 Query: 543 NKFEES----QIKYKGDASEISN*NM-------G*RKNNGVWSV*EQVPT*NISHPNLLL 689 +K+E ++ +AS I + + G RK + + P L+ Sbjct: 358 SKYEPRFHVMDVQGSTEASAIKDYVVKHALPLVGHRKTSNDAKRYSKRP---------LV 408 Query: 690 VALYSVDYEKNPKG-TNYWRNRIMKVAKKFVDSGKSVHFAISNSQEQSHELSEFGIESPD 866 V YSVD+ + + T +WRN++++VAK F + FAI++ ++ + E+ + G+ S Sbjct: 409 VVYYSVDFSFDYRAATQFWRNKVLEVAKDFPE----YTFAIADEEDYATEVKDLGL-SES 463 Query: 867 SNKVYIVARDEKNQKFKMA-EDFTMEAFEKFINDFLNEQVPVYLKSETIPEKNDEPVKVV 1043 V DE +KF M E+F + +F+ F ++ +KS+ +P+ N PVKVV Sbjct: 464 GEDVNAAILDESGKKFAMEPEEFDSDTLREFVTAFKKGKLKPVIKSQPVPKNNKGPVKVV 523 Query: 1044 VAKNFDALVNDPTKDVLIEFYAPWCGHCKSLAPKYEELAKKLSGESDIVIAKMDATANDV 1223 V K FDA+V DP KDVLIEFYAPWCGHCK L P Y L KK G+ D+VIAKMDATAND+ Sbjct: 524 VGKTFDAIVMDPKKDVLIEFYAPWCGHCKQLEPIYTSLGKKYKGQKDLVIAKMDATANDI 583 Query: 1224 PS-PYEVTGFPTIYFAPKNSKNSPKSYNGG-REVDDMLKYIASEST 1355 + Y+V GFPTIYFAP K +P + GG R+++ + K+I +T Sbjct: 584 TNDQYKVEGFPTIYFAPSGDKKNPIKFEGGNRDLEHLSKFIDEHAT 629
Score = 119 bits (297), Expect = 3e-26
Identities = 60/134 (44%), Positives = 83/134 (61%), Gaps = 6/134 (4%)
Frame = +3
Query: 6 NFEQSIKEHSVILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETKTCE 185
NF+ + + +L+EFYAPWCGHCK+ APEYE A+TLK DPPI +AK+D T+ +
Sbjct: 64 NFDNFVADKDTVLLEFYAPWCGHCKQFAPEYEKIASTLKDNDPPIAVAKIDATSASMLAS 123
Query: 186 KHGVSGYPTLKIFKNGQFAFDYSGERSSDGIIKYMISKSGPS---SKEVK---TEENFKK 347
K VSGYPT+KI K GQ A DY G R+ + I+ + S P EV T++NF
Sbjct: 124 KFDVSGYPTIKILKKGQ-AVDYDGSRTQEEIVAKVREVSQPDWTPPPEVTLSLTKDNFDD 182
Query: 348 FIASTDPVVVGFFA 389
+ + D ++V F+A
Sbjct: 183 VVNNADIILVEFYA 196
Score = 78.2 bits (191), Expect = 6e-14
Identities = 47/115 (40%), Positives = 68/115 (59%), Gaps = 3/115 (2%)
Frame = +3
Query: 1017 KNDEPVKVVVAKNFDALVNDPTKD-VLIEFYAPWCGHCKSLAPKYEELAKKL-SGESDIV 1190
K + V V+ NFD V D KD VL+EFYAPWCGHCK AP+YE++A L + I
Sbjct: 52 KEENGVWVLNDGNFDNFVAD--KDTVLLEFYAPWCGHCKQFAPEYEKIASTLKDNDPPIA 109
Query: 1191 IAKMDAT-ANDVPSPYEVTGFPTIYFAPKNSKNSPKSYNGGREVDDMLKYIASES 1352
+AK+DAT A+ + S ++V+G+PTI K Y+G R ++++ + S
Sbjct: 110 VAKIDATSASMLASKFDVSGYPTIKIL---KKGQAVDYDGSRTQEEIVAKVREVS 161
>sp|P38659|PDIA4_RAT Protein disulfide-isomerase A4 precursor (Protein ERp-72) (ERp72) (Calcium-binding protein 2) (CaBP2) Length = 643 Score = 340 bits (872), Expect = 7e-93 Identities = 190/466 (40%), Positives = 271/466 (58%), Gaps = 16/466 (3%) Frame = +3 Query: 6 NFEQSIKEHSVILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETKTCE 185 NF+ + +ILVEFYAPWCGHCKKLAPEYE AA L ++ PPIP+AKVD T +T + Sbjct: 184 NFDDVVNNADIILVEFYAPWCGHCKKLAPEYEKAAKELSKRSPPIPLAKVDATEQTDLAK 243 Query: 186 KHGVSGYPTLKIFKNGQFAFDYSGERSSDGIIKYMISKSGPSSKEVKTEENFKKFIASTD 365 + VSGYPTLKIF+ G+ FDY+G R GI+ YM+ +SGP SKE+ T + ++F+ D Sbjct: 244 RFDVSGYPTLKIFRKGR-PFDYNGpreKYGIVDYMVEQSGPPSKEILTLKQVQEFLKDGD 302 Query: 366 PVVV-GFFASESSDLYKAFLSVAEKLNEDFRVAHVFDESFESKIKFRKTTFVIYHPQKMA 542 VV+ G F Y + A L ED++ H F +K V+ P+K Sbjct: 303 DVVILGVFQGVGDPGYLQYQDAANTLREDYKFHHTFSTEIAKFLKVSLGKLVLMQPEKFQ 362 Query: 543 NKFEESQ----IKYKGDASEISN*NM-------G*RKNNGVWSV*EQVPT*NISHPNLLL 689 +K+E ++ +AS I + + G RK + + P L+ Sbjct: 363 SKYEPRMHVMDVQGSTEASAIKDYVVKHALPLVGHRKTSNDAKRYSKRP---------LV 413 Query: 690 VALYSVDYEKNPK-GTNYWRNRIMKVAKKFVDSGKSVHFAISNSQEQSHELSEFGIESPD 866 V YSVD+ + + T +WRN++++VAK F + FAI++ ++ + E+ + G+ S Sbjct: 414 VVYYSVDFSFDYRTATQFWRNKVLEVAKDFPE----YTFAIADEEDYATEVKDLGL-SES 468 Query: 867 SNKVYIVARDEKNQKFKMA-EDFTMEAFEKFINDFLNEQVPVYLKSETIPEKNDEPVKVV 1043 V DE +KF M E+F +A ++F+ F ++ +KS+ +P+ N PV+VV Sbjct: 469 GEDVNAAILDESGKKFAMEPEEFDSDALQEFVMAFKKGKLKPVIKSQPVPKNNKGPVRVV 528 Query: 1044 VAKNFDALVNDPTKDVLIEFYAPWCGHCKSLAPKYEELAKKLSGESDIVIAKMDATANDV 1223 V K FDA+V DP KDVLIEFYAPWCGHCK L P Y L KK G+ D+VIAKMDATAND+ Sbjct: 529 VGKTFDAIVMDPKKDVLIEFYAPWCGHCKQLEPVYTSLGKKYKGQKDLVIAKMDATANDI 588 Query: 1224 PSP-YEVTGFPTIYFAPKNSKNSPKSYNGG-REVDDMLKYIASEST 1355 + Y+V GFPTIYFAP K +P + GG R+++ + K+I +T Sbjct: 589 TNDRYKVEGFPTIYFAPSGDKKNPIKFEGGNRDLEHLSKFIDEHAT 634
Score = 120 bits (300), Expect = 1e-26
Identities = 61/134 (45%), Positives = 83/134 (61%), Gaps = 6/134 (4%)
Frame = +3
Query: 6 NFEQSIKEHSVILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETKTCE 185
NF+ + + +L+EFYAPWCGHCK+ APEYE A+TLK DPPI +AK+D T+ +
Sbjct: 69 NFDNFVADKDTVLLEFYAPWCGHCKQFAPEYEKIASTLKDNDPPIAVAKIDATSASMLAS 128
Query: 186 KHGVSGYPTLKIFKNGQFAFDYSGERSSDGIIKYMISKSGPS---SKEVK---TEENFKK 347
K VSGYPT+KI K GQ A DY G R+ + I+ + S P EV T+ENF
Sbjct: 129 KFDVSGYPTIKILKKGQ-AVDYDGSRTQEEIVAKVREVSQPDWTPPPEVTLTLTKENFDD 187
Query: 348 FIASTDPVVVGFFA 389
+ + D ++V F+A
Sbjct: 188 VVNNADIILVEFYA 201
Score = 87.0 bits (214), Expect = 1e-16
Identities = 48/111 (43%), Positives = 69/111 (62%), Gaps = 2/111 (1%)
Frame = +3
Query: 1026 EPVKVVVAKNFDALVNDPTKDVLIEFYAPWCGHCKSLAPKYEELAKKLSGES-DIVIAKM 1202
E + +NFD +VN+ +L+EFYAPWCGHCK LAP+YE+ AK+LS S I +AK+
Sbjct: 175 EVTLTLTKENFDDVVNN-ADIILVEFYAPWCGHCKKLAPEYEKAAKELSKRSPPIPLAKV 233
Query: 1203 DAT-ANDVPSPYEVTGFPTIYFAPKNSKNSPKSYNGGREVDDMLKYIASES 1352
DAT D+ ++V+G+PT+ K P YNG RE ++ Y+ +S
Sbjct: 234 DATEQTDLAKRFDVSGYPTLKIF---RKGRPFDYNGpreKYGIVDYMVEQS 281
Score = 79.3 bits (194), Expect = 3e-14
Identities = 47/115 (40%), Positives = 69/115 (60%), Gaps = 3/115 (2%)
Frame = +3
Query: 1017 KNDEPVKVVVAKNFDALVNDPTKD-VLIEFYAPWCGHCKSLAPKYEELAKKL-SGESDIV 1190
K + V V+ +NFD V D KD VL+EFYAPWCGHCK AP+YE++A L + I
Sbjct: 57 KEENGVWVLNDENFDNFVAD--KDTVLLEFYAPWCGHCKQFAPEYEKIASTLKDNDPPIA 114
Query: 1191 IAKMDAT-ANDVPSPYEVTGFPTIYFAPKNSKNSPKSYNGGREVDDMLKYIASES 1352
+AK+DAT A+ + S ++V+G+PTI K Y+G R ++++ + S
Sbjct: 115 VAKIDATSASMLASKFDVSGYPTIKIL---KKGQAVDYDGSRTQEEIVAKVREVS 166
>sp|P34329|PDIA4_CAEEL Probable protein disulfide-isomerase A4 precursor (ERp-72 homolog) Length = 618 Score = 324 bits (831), Expect = 4e-88 Identities = 182/469 (38%), Positives = 273/469 (58%), Gaps = 12/469 (2%) Frame = +3 Query: 6 NFEQSIKEHSVILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETKTCE 185 NF+ I + ++LVEFYAPWCGHCKKLAPEYE AA LK Q + + KVD T E Sbjct: 156 NFDDFISNNELVLVEFYAPWCGHCKKLAPEYEKAAQKLKAQGSKVKLGKVDATIEKDLGT 215 Query: 186 KHGVSGYPTLKIFKNGQFAFDYSGERSSDGIIKYMISKSGPSSKEVKTEENFKKFIASTD 365 K+GVSGYPT+KI +NG+ FDY+G R + GIIKYM +S P++K++ ++ ++F++ D Sbjct: 216 KYGVSGYPTMKIIRNGR-RFDYNGpreAAGIIKYMTDQSKPAAKKLPKLKDVERFMSKDD 274 Query: 366 PVVVGFFASESSDLYKAFLSVAEKLNEDFR-VAHVFDESFESKIKFRKTTFVIYHPQKMA 542 ++GFFA+E S ++AF AE L E+F+ + H D + K + +I++P Sbjct: 275 VTIIGFFATEDSTAFEAFSDSAEMLREEFKTMGHTSDPAAFKKWDAKPNDIIIFYPSLFH 334 Query: 543 NKFEESQIKYKGDASEISN*NMG*RKNNGVW---SV*EQVPT*NISHPNLLLVALYSVDY 713 +KFE Y A+ + R+++ + T P L+V Y+ D+ Sbjct: 335 SKFEPKSRTYNKAAATSEDLLAFFREHSAPLVGKMTKKNAATRYTKKP--LVVVYYNADF 392 Query: 714 E-KNPKGTNYWRNRIMKVAKKFVDSGKSVHFAISNSQEQSHELSEFGI-ESPDSNKVYIV 887 + +G+ YWR++++ +A+K+ FA+++ +E + EL E G+ +S + V + Sbjct: 393 SVQYREGSEYWRSKVLNIAQKY--QKDKYKFAVADEEEFAKELEELGLGDSGLEHNVVVF 450 Query: 888 ARDEKN-----QKFKMAEDFTMEAFEKFINDFLNEQVPVYLKSETIPEKNDEPVKVVVAK 1052 D K +F D +EAF K I+ + + ++KS P+ + PVK VV Sbjct: 451 GYDGKKYPMNPDEFDGELDENLEAFMKQIS---SGKAKAHVKSAPAPKDDKGPVKTVVGS 507 Query: 1053 NFDALVNDPTKDVLIEFYAPWCGHCKSLAPKYEELAKKL-SGESDIVIAKMDATANDVPS 1229 NFD +VND +KDVLIEFYAPWCGHCKS KY ELA+ L + ++V+AKMDAT ND PS Sbjct: 508 NFDKIVNDESKDVLIEFYAPWCGHCKSFESKYVELAQALKKTQPNVVLAKMDATINDAPS 567 Query: 1230 PYEVTGFPTIYFAPKNSKNSPKSYNGGREVDDMLKYIASESTSSLKSYD 1376 + V GFPTIYFAP K+ P Y+G R+++D+ K++ S + D Sbjct: 568 QFAVEGFPTIYFAPAGKKSEPIKYSGNRDLEDLKKFMTKHGVKSFQKKD 616
Score = 114 bits (285), Expect = 8e-25
Identities = 58/134 (43%), Positives = 86/134 (64%), Gaps = 6/134 (4%)
Frame = +3
Query: 6 NFEQSIKEHSVILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETKTCE 185
NF+ +K++ +LV+FYAPWCGHCK LAPEYE A++ + IP+AKVD T ET+ +
Sbjct: 45 NFDAFLKKNPSVLVKFYAPWCGHCKHLAPEYEKASSKVS-----IPLAKVDATVETELGK 99
Query: 186 KHGVSGYPTLKIFKNGQFAFDYSGERSSDGIIKYMISKSGPSSKE------VKTEENFKK 347
+ + GYPTLK +K+G+ DY G R GI++++ S+ P+ K T ENF
Sbjct: 100 RFEIQGYPTLKFWKDGKGPNDYDGGRDEAGIVEWVESRVDPNYKPPPEEVVTLTTENFDD 159
Query: 348 FIASTDPVVVGFFA 389
FI++ + V+V F+A
Sbjct: 160 FISNNELVLVEFYA 173
Score = 86.7 bits (213), Expect = 2e-16
Identities = 48/116 (41%), Positives = 73/116 (62%), Gaps = 1/116 (0%)
Frame = +3
Query: 1023 DEPVKVVVAKNFDALVNDPTKDVLIEFYAPWCGHCKSLAPKYEELAKKLSGESDIVIAKM 1202
DE V V+ KNFDA + VL++FYAPWCGHCK LAP+YE+ + K+S I +AK+
Sbjct: 35 DEGVVVLTDKNFDAFLKK-NPSVLVKFYAPWCGHCKHLAPEYEKASSKVS----IPLAKV 89
Query: 1203 DATA-NDVPSPYEVTGFPTIYFAPKNSKNSPKSYNGGREVDDMLKYIASESTSSLK 1367
DAT ++ +E+ G+PT+ F K+ K P Y+GGR+ +++++ S + K
Sbjct: 90 DATVETELGKRFEIQGYPTLKFW-KDGK-GPNDYDGGRDEAGIVEWVESRVDPNYK 143
>sp|P55059|PDI_HUMIN Protein disulfide-isomerase precursor (PDI) Length = 505 Score = 274 bits (700), Expect = 6e-73 Identities = 169/458 (36%), Positives = 249/458 (54%), Gaps = 13/458 (2%) Frame = +3 Query: 9 FEQSIKEHSVILVEFYAPWCGHCKKLAPEYEIAATTLKRQDPPIPIAKVDCTAETKTCEK 188 F+ IK + ++L EF+APWCGHCK LAPEYE AATTLK ++ I +AKVDCT ET C++ Sbjct: 31 FDDFIKTNDLVLAEFFAPWCGHCKALAPEYEEAATTLKEKN--IKLAKVDCTEETDLCQQ 88 Query: 189 HGVSGYPTLKIFKNGQFAFDYSGERSSDGIIKYMISKSGPSSKEVKTEENFKKFIASTDP 368 HGV GYPTLK+F+ Y G+R + I YMI +S P+ EV T++N ++F + Sbjct: 89 HGVEGYPTLKVFRGLDNVSPYKGQRKAAAITSYMIKQSLPAVSEV-TKDNLEEFKKADKA 147 Query: 369 VVVGFFASESSDLYKAFLSVAEKLNEDFRVAHVFDESFESKIKFRKTTFVIYHP-----Q 533 V+V + + + F VAEKL +++ D + + V+Y Sbjct: 148 VLVAYVDASDKASSEVFTQVAEKLRDNYPFGSSSDAALAEAEGVKAPAIVLYKDFDEGKA 207 Query: 534 KMANKFEESQIKY--KGDASEISN*NMG*RKNNGVWSV*EQVPT*NISHPNLLLVALYSV 707 + KFE I+ K A+ + +G + S + L +++ Sbjct: 208 VFSEKFEVEAIEKFAKTGATPLIG-EIGPETYSDYMSA------------GIPLAYIFAE 254 Query: 708 DYEKNPKGTNYWRNRIMKVAKKFVDSGKSV-HFAISNSQEQSHELSEFGIESPDSNKVYI 884 E+ R + K ++ + V +F +++ +++ D + Sbjct: 255 TAEE--------RKELSDKLKPIAEAQRGVINFGTIDAKAFGAHAGNLNLKT-DKFPAFA 305 Query: 885 VARDEKNQKFKMAED--FTMEAFEKFINDFLNEQVPVYLKSETIPEKNDEPVKVVVAKNF 1058 + KNQKF ++ T EA + F++DF+ ++ +KSE IPEK + PV VVVAKN+ Sbjct: 306 IQEVAKNQKFPFDQEKEITFEAIKAFVDDFVAGKIEPSIKSEPIPEKQEGPVTVVVAKNY 365 Query: 1059 DALVNDPTKDVLIEFYAPWCGHCKSLAPKYEELA---KKLSGESDIVIAKMDATANDVPS 1229 + +V D TKDVLIEFYAPWCGHCK+LAPKYEEL K + +VIAK+DATANDVP Sbjct: 366 NEIVLDDTKDVLIEFYAPWCGHCKALAPKYEELGALYAKSEFKDRVVIAKVDATANDVPD 425 Query: 1230 PYEVTGFPTIYFAPKNSKNSPKSYNGGREVDDMLKYIA 1343 E+ GFPTI P +K P +Y+G R V+D++K+IA Sbjct: 426 --EIQGFPTIKLYPAGAKGQPVTYSGSRTVEDLIKFIA 461
Score = 68.2 bits (165), Expect = 6e-11
Identities = 44/117 (37%), Positives = 64/117 (54%), Gaps = 3/117 (2%)
Frame = +3
Query: 1089 VLIEFYAPWCGHCKSLAPKYEELAKKLSGESDIVIAKMDAT-ANDVPSPYEVTGFPTIYF 1265
VL EF+APWCGHCK+LAP+YEE A L E +I +AK+D T D+ + V G+PT+
Sbjct: 41 VLAEFFAPWCGHCKALAPEYEEAATTLK-EKNIKLAKVDCTEETDLCQQHGVEGYPTLKV 99
Query: 1266 APKNSKNSPKSYNGGREVDDMLKYIASESTSSLK--SYDRQGRVRKSDL*KIKIVEY 1430
SP Y G R+ + Y+ +S ++ + D +K+D K +V Y
Sbjct: 100 FRGLDNVSP--YKGQRKAAAITSYMIKQSLPAVSEVTKDNLEEFKKAD--KAVLVAY 152
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 167,359,106
Number of Sequences: 369166
Number of extensions: 3428369
Number of successful extensions: 10830
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9441
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10468
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 18608629725
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail