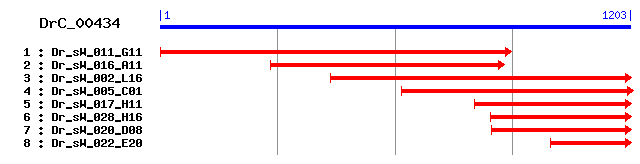
DrC_00434
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00434 (1202 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q90835|EF1A_CHICK Elongation factor 1-alpha 1 (EF-1-alph... 608 e-174 sp|P10126|EF1A1_MOUSE Elongation factor 1-alpha 1 (EF-1-alp... 607 e-173 sp|P68105|EF1A1_RABIT Elongation factor 1-alpha 1 (EF-1-alp... 607 e-173 sp|P13549|EF1A0_XENLA Elongation factor 1-alpha, somatic fo... 606 e-173 sp|P27592|EF1A_ONCVO Elongation factor 1-alpha (EF-1-alpha) 606 e-173 sp|Q92005|EF1A_BRARE Elongation factor 1-alpha (EF-1-alpha) 606 e-173 sp|Q5R4R8|EF1A1_PONPY Elongation factor 1-alpha 1 (EF-1-alp... 605 e-173 sp|Q71V39|EF1A2_RABIT Elongation factor 1-alpha 2 (EF-1-alp... 603 e-172 sp|P62631|EF1A2_MOUSE Elongation factor 1-alpha 2 (EF-1-alp... 602 e-172 sp|P17508|EF1A3_XENLA Elongation factor 1-alpha, oocyte for... 601 e-171
>sp|Q90835|EF1A_CHICK Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor Tu) (EF-Tu) Length = 462 Score = 608 bits (1569), Expect = e-174 Identities = 291/368 (79%), Positives = 330/368 (89%) Frame = +1 Query: 1 ALWKFETTKFYVTVIDAPGHRDFIKNMITGTSQADCAVLVVAAGTGEFEAGISKNGQTRE 180 +LWKFET+K+YVT+IDAPGHRDFIKNMITGTSQADCAVL+VAAG GEFEAGISKNGQTRE Sbjct: 76 SLWKFETSKYYVTIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAGISKNGQTRE 135 Query: 181 HALLAYTLGVKQMIVGVNKMDSTEPPFSESRFNEIRKEVSSYLKKVGYQPDGVAFVPISG 360 HALLAYTLGVKQ+IVGVNKMDSTEPP+S+ R+ EI KEVS+Y+KK+GY PD VAFVPISG Sbjct: 136 HALLAYTLGVKQLIVGVNKMDSTEPPYSQKRYEEIVKEVSTYIKKIGYNPDTVAFVPISG 195 Query: 361 WNGDNMIEESSNMSWYKGWDITRKNAKKEETKITGKTLLDALDSLEPPSRPTNKPLRLPL 540 WNGDNM+E SSNM W+KGW +TRK+ +G TLL+ALD + PP+RPT+KPLRLPL Sbjct: 196 WNGDNMLEPSSNMPWFKGWKVTRKDGNA-----SGTTLLEALDCILPPTRPTDKPLRLPL 250 Query: 541 QDVYKIGGIGTVPVGRVETGILKPGMVVTFAPQGLTTEVKSVEMHHEALTEALPGDNVGF 720 QDVYKIGGIGTVPVGRVETG+LKPGMVVTFAP +TTEVKSVEMHHEAL+EALPGDNVGF Sbjct: 251 QDVYKIGGIGTVPVGRVETGVLKPGMVVTFAPVNVTTEVKSVEMHHEALSEALPGDNVGF 310 Query: 721 NIKNVSVKEIRRGNVCSDSKSDPARQATDFIAQVIIMNHPGEIHSGYSPVLDCHTAHIAC 900 N+KNVSVK++RRGNV DSK+DP +A F AQVII+NHPG+I +GY+PVLDCHTAHIAC Sbjct: 311 NVKNVSVKDVRRGNVAGDSKNDPPMEAAGFTAQVIILNHPGQISAGYAPVLDCHTAHIAC 370 Query: 901 KFAELLKKIDRRSGKDLEDFPKQFKSGDAGLVKLIPSKPMCVETFSEYPPLGRFAVRDMK 1080 KFAEL +KIDRRSGK LED PK KSGDA +V +IP KPMCVE+FS+YPPLGRFAVRDM+ Sbjct: 371 KFAELKEKIDRRSGKKLEDGPKFLKSGDAAIVDMIPGKPMCVESFSDYPPLGRFAVRDMR 430 Query: 1081 QTVAVGVI 1104 QTVAVGVI Sbjct: 431 QTVAVGVI 438
>sp|P10126|EF1A1_MOUSE Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu) sp|P62630|EF1A1_RAT Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu) sp|P62629|EF1A1_CRIGR Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu) Length = 462 Score = 607 bits (1565), Expect = e-173 Identities = 289/368 (78%), Positives = 330/368 (89%) Frame = +1 Query: 1 ALWKFETTKFYVTVIDAPGHRDFIKNMITGTSQADCAVLVVAAGTGEFEAGISKNGQTRE 180 +LWKFET+K+YVT+IDAPGHRDFIKNMITGTSQADCAVL+VAAG GEFEAGISKNGQTRE Sbjct: 76 SLWKFETSKYYVTIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAGISKNGQTRE 135 Query: 181 HALLAYTLGVKQMIVGVNKMDSTEPPFSESRFNEIRKEVSSYLKKVGYQPDGVAFVPISG 360 HALLAYTLGVKQ+IVGVNKMDSTEPP+S+ R+ EI KEVS+Y+KK+GY PD VAFVPISG Sbjct: 136 HALLAYTLGVKQLIVGVNKMDSTEPPYSQKRYEEIVKEVSTYIKKIGYNPDTVAFVPISG 195 Query: 361 WNGDNMIEESSNMSWYKGWDITRKNAKKEETKITGKTLLDALDSLEPPSRPTNKPLRLPL 540 WNGDNM+E S+NM W+KGW +TRK+ +G TLL+ALD + PP+RPT+KPLRLPL Sbjct: 196 WNGDNMLEPSANMPWFKGWKVTRKDGSA-----SGTTLLEALDCILPPTRPTDKPLRLPL 250 Query: 541 QDVYKIGGIGTVPVGRVETGILKPGMVVTFAPQGLTTEVKSVEMHHEALTEALPGDNVGF 720 QDVYKIGGIGTVPVGRVETG+LKPGMVVTFAP +TTEVKSVEMHHEAL+EALPGDNVGF Sbjct: 251 QDVYKIGGIGTVPVGRVETGVLKPGMVVTFAPVNVTTEVKSVEMHHEALSEALPGDNVGF 310 Query: 721 NIKNVSVKEIRRGNVCSDSKSDPARQATDFIAQVIIMNHPGEIHSGYSPVLDCHTAHIAC 900 N+KNVSVK++RRGNV DSK+DP +A F AQVII+NHPG+I +GY+PVLDCHTAHIAC Sbjct: 311 NVKNVSVKDVRRGNVAGDSKNDPPMEAAGFTAQVIILNHPGQISAGYAPVLDCHTAHIAC 370 Query: 901 KFAELLKKIDRRSGKDLEDFPKQFKSGDAGLVKLIPSKPMCVETFSEYPPLGRFAVRDMK 1080 KFAEL +KIDRRSGK LED PK KSGDA +V ++P KPMCVE+FS+YPPLGRFAVRDM+ Sbjct: 371 KFAELKEKIDRRSGKKLEDGPKFLKSGDAAIVDMVPGKPMCVESFSDYPPLGRFAVRDMR 430 Query: 1081 QTVAVGVI 1104 QTVAVGVI Sbjct: 431 QTVAVGVI 438
>sp|P68105|EF1A1_RABIT Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu) sp|Q5R1X2|EF1A1_PANTR Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu) sp|P68103|EF1A1_BOVIN Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu) sp|P68104|EF1A1_HUMAN Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu) sp|Q66RN5|EF1A1_FELCA Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu) Length = 462 Score = 607 bits (1565), Expect = e-173 Identities = 289/368 (78%), Positives = 330/368 (89%) Frame = +1 Query: 1 ALWKFETTKFYVTVIDAPGHRDFIKNMITGTSQADCAVLVVAAGTGEFEAGISKNGQTRE 180 +LWKFET+K+YVT+IDAPGHRDFIKNMITGTSQADCAVL+VAAG GEFEAGISKNGQTRE Sbjct: 76 SLWKFETSKYYVTIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAGISKNGQTRE 135 Query: 181 HALLAYTLGVKQMIVGVNKMDSTEPPFSESRFNEIRKEVSSYLKKVGYQPDGVAFVPISG 360 HALLAYTLGVKQ+IVGVNKMDSTEPP+S+ R+ EI KEVS+Y+KK+GY PD VAFVPISG Sbjct: 136 HALLAYTLGVKQLIVGVNKMDSTEPPYSQKRYEEIVKEVSTYIKKIGYNPDTVAFVPISG 195 Query: 361 WNGDNMIEESSNMSWYKGWDITRKNAKKEETKITGKTLLDALDSLEPPSRPTNKPLRLPL 540 WNGDNM+E S+NM W+KGW +TRK+ +G TLL+ALD + PP+RPT+KPLRLPL Sbjct: 196 WNGDNMLEPSANMPWFKGWKVTRKDGNA-----SGTTLLEALDCILPPTRPTDKPLRLPL 250 Query: 541 QDVYKIGGIGTVPVGRVETGILKPGMVVTFAPQGLTTEVKSVEMHHEALTEALPGDNVGF 720 QDVYKIGGIGTVPVGRVETG+LKPGMVVTFAP +TTEVKSVEMHHEAL+EALPGDNVGF Sbjct: 251 QDVYKIGGIGTVPVGRVETGVLKPGMVVTFAPVNVTTEVKSVEMHHEALSEALPGDNVGF 310 Query: 721 NIKNVSVKEIRRGNVCSDSKSDPARQATDFIAQVIIMNHPGEIHSGYSPVLDCHTAHIAC 900 N+KNVSVK++RRGNV DSK+DP +A F AQVII+NHPG+I +GY+PVLDCHTAHIAC Sbjct: 311 NVKNVSVKDVRRGNVAGDSKNDPPMEAAGFTAQVIILNHPGQISAGYAPVLDCHTAHIAC 370 Query: 901 KFAELLKKIDRRSGKDLEDFPKQFKSGDAGLVKLIPSKPMCVETFSEYPPLGRFAVRDMK 1080 KFAEL +KIDRRSGK LED PK KSGDA +V ++P KPMCVE+FS+YPPLGRFAVRDM+ Sbjct: 371 KFAELKEKIDRRSGKKLEDGPKFLKSGDAAIVDMVPGKPMCVESFSDYPPLGRFAVRDMR 430 Query: 1081 QTVAVGVI 1104 QTVAVGVI Sbjct: 431 QTVAVGVI 438
>sp|P13549|EF1A0_XENLA Elongation factor 1-alpha, somatic form (EF-1-alpha-S) Length = 462 Score = 606 bits (1563), Expect = e-173 Identities = 291/368 (79%), Positives = 329/368 (89%) Frame = +1 Query: 1 ALWKFETTKFYVTVIDAPGHRDFIKNMITGTSQADCAVLVVAAGTGEFEAGISKNGQTRE 180 +LWKFET+K+YVT+IDAPGHRDFIKNMITGTSQADCAVL+VAAG GEFEAGISKNGQTRE Sbjct: 76 SLWKFETSKYYVTIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAGISKNGQTRE 135 Query: 181 HALLAYTLGVKQMIVGVNKMDSTEPPFSESRFNEIRKEVSSYLKKVGYQPDGVAFVPISG 360 HALLAYTLGVKQ+IVG+NKMDSTEPP+S+ R+ EI KEVS+Y+KK+GY PD VAFVPISG Sbjct: 136 HALLAYTLGVKQLIVGINKMDSTEPPYSQKRYEEIVKEVSTYIKKIGYNPDTVAFVPISG 195 Query: 361 WNGDNMIEESSNMSWYKGWDITRKNAKKEETKITGKTLLDALDSLEPPSRPTNKPLRLPL 540 WNGDNM+E S NM W+KGW ITRK E +G TLL+ALD + PPSRPT+KPLRLPL Sbjct: 196 WNGDNMLEPSPNMPWFKGWKITRK-----EGSGSGTTLLEALDCILPPSRPTDKPLRLPL 250 Query: 541 QDVYKIGGIGTVPVGRVETGILKPGMVVTFAPQGLTTEVKSVEMHHEALTEALPGDNVGF 720 QDVYKIGGIGTVPVGRVETG++KPGMVVTFAP +TTEVKSVEMHHEALTEA+PGDNVGF Sbjct: 251 QDVYKIGGIGTVPVGRVETGVIKPGMVVTFAPVNVTTEVKSVEMHHEALTEAVPGDNVGF 310 Query: 721 NIKNVSVKEIRRGNVCSDSKSDPARQATDFIAQVIIMNHPGEIHSGYSPVLDCHTAHIAC 900 N+KNVSVK++RRGNV DSK+DP +A F AQVII+NHPG+I +GY+PVLDCHTAHIAC Sbjct: 311 NVKNVSVKDVRRGNVAGDSKNDPPMEAGSFTAQVIILNHPGQIGAGYAPVLDCHTAHIAC 370 Query: 901 KFAELLKKIDRRSGKDLEDFPKQFKSGDAGLVKLIPSKPMCVETFSEYPPLGRFAVRDMK 1080 KFAEL +KIDRRSGK LED PK KSGDA +V +IP KPMCVE+FS+YPPLGRFAVRDM+ Sbjct: 371 KFAELKEKIDRRSGKKLEDNPKFLKSGDAAIVDMIPGKPMCVESFSDYPPLGRFAVRDMR 430 Query: 1081 QTVAVGVI 1104 QTVAVGVI Sbjct: 431 QTVAVGVI 438
>sp|P27592|EF1A_ONCVO Elongation factor 1-alpha (EF-1-alpha) Length = 464 Score = 606 bits (1563), Expect = e-173 Identities = 289/368 (78%), Positives = 329/368 (89%) Frame = +1 Query: 1 ALWKFETTKFYVTVIDAPGHRDFIKNMITGTSQADCAVLVVAAGTGEFEAGISKNGQTRE 180 ALWKFET K+Y+T+IDAPGHRDFIKNMITGTSQADCAVLVVA GTGEFEAGISKNGQTRE Sbjct: 76 ALWKFETPKYYITIIDAPGHRDFIKNMITGTSQADCAVLVVACGTGEFEAGISKNGQTRE 135 Query: 181 HALLAYTLGVKQMIVGVNKMDSTEPPFSESRFNEIRKEVSSYLKKVGYQPDGVAFVPISG 360 HALLA TLGVKQMIV NKMDST+PPFSE+RF E+ EVS+Y+KK+GY P + FVPISG Sbjct: 136 HALLAQTLGVKQMIVACNKMDSTDPPFSEARFGEVTTEVSNYIKKIGYNPKSIPFVPISG 195 Query: 361 WNGDNMIEESSNMSWYKGWDITRKNAKKEETKITGKTLLDALDSLEPPSRPTNKPLRLPL 540 +NGDNM+E S+NM W+KGW + RK E +TGKTLL+ALDS+ PP RPT+KPLRLPL Sbjct: 196 FNGDNMLEPSANMPWFKGWSVERK-----EGTMTGKTLLEALDSVVPPQRPTDKPLRLPL 250 Query: 541 QDVYKIGGIGTVPVGRVETGILKPGMVVTFAPQGLTTEVKSVEMHHEALTEALPGDNVGF 720 QDVYKIGGIGTVPVGRVETGILKPGM+VTFAPQ LTTEVKSVEMHHEAL EALPGDNVGF Sbjct: 251 QDVYKIGGIGTVPVGRVETGILKPGMIVTFAPQNLTTEVKSVEMHHEALQEALPGDNVGF 310 Query: 721 NIKNVSVKEIRRGNVCSDSKSDPARQATDFIAQVIIMNHPGEIHSGYSPVLDCHTAHIAC 900 N+KN+S+K+IRRG+V SDSK+DPA++ F AQVIIMNHPG+I +GY+PVLDCHTAHIAC Sbjct: 311 NVKNISIKDIRRGSVASDSKNDPAKETKMFTAQVIIMNHPGQISAGYTPVLDCHTAHIAC 370 Query: 901 KFAELLKKIDRRSGKDLEDFPKQFKSGDAGLVKLIPSKPMCVETFSEYPPLGRFAVRDMK 1080 KFAEL +K+DRRSGK +ED PK KSGDAG++ LIP+KP+CVETF+EYPPLGRFAVRDM+ Sbjct: 371 KFAELKEKVDRRSGKKVEDNPKSLKSGDAGIIDLIPTKPLCVETFTEYPPLGRFAVRDMR 430 Query: 1081 QTVAVGVI 1104 QTVAVGVI Sbjct: 431 QTVAVGVI 438
>sp|Q92005|EF1A_BRARE Elongation factor 1-alpha (EF-1-alpha) Length = 462 Score = 606 bits (1563), Expect = e-173 Identities = 291/368 (79%), Positives = 328/368 (89%) Frame = +1 Query: 1 ALWKFETTKFYVTVIDAPGHRDFIKNMITGTSQADCAVLVVAAGTGEFEAGISKNGQTRE 180 ALWKFET+K+YVT+IDAPGHRDFIKNMITGTSQADCAVL+VA G GEFEAGISKNGQTRE Sbjct: 76 ALWKFETSKYYVTIIDAPGHRDFIKNMITGTSQADCAVLIVAGGVGEFEAGISKNGQTRE 135 Query: 181 HALLAYTLGVKQMIVGVNKMDSTEPPFSESRFNEIRKEVSSYLKKVGYQPDGVAFVPISG 360 HALLA+TLGVKQ+IVGVNKMDSTEPP+S++RF EI KEVS+Y+KK+GY P VAFVPISG Sbjct: 136 HALLAFTLGVKQLIVGVNKMDSTEPPYSQARFEEITKEVSAYIKKIGYNPASVAFVPISG 195 Query: 361 WNGDNMIEESSNMSWYKGWDITRKNAKKEETKITGKTLLDALDSLEPPSRPTNKPLRLPL 540 W+GDNM+E SSNM W+KGW I RK E +G TLLDALD++ PPSRPT+KPLRLPL Sbjct: 196 WHGDNMLEASSNMGWFKGWKIERK-----EGNASGTTLLDALDAILPPSRPTDKPLRLPL 250 Query: 541 QDVYKIGGIGTVPVGRVETGILKPGMVVTFAPQGLTTEVKSVEMHHEALTEALPGDNVGF 720 QDVYKIGGIGTVPVGRVETG+LKPGMVVTFAP +TTEVKSVEMHHE+LTEA PGDNVGF Sbjct: 251 QDVYKIGGIGTVPVGRVETGVLKPGMVVTFAPANVTTEVKSVEMHHESLTEATPGDNVGF 310 Query: 721 NIKNVSVKEIRRGNVCSDSKSDPARQATDFIAQVIIMNHPGEIHSGYSPVLDCHTAHIAC 900 N+KNVSVK+IRRGNV DSK+DP +A +F AQVII+NHPG+I GY+PVLDCHTAHIAC Sbjct: 311 NVKNVSVKDIRRGNVAGDSKNDPPMEAANFNAQVIILNHPGQISQGYAPVLDCHTAHIAC 370 Query: 901 KFAELLKKIDRRSGKDLEDFPKQFKSGDAGLVKLIPSKPMCVETFSEYPPLGRFAVRDMK 1080 KFAEL +KIDRRSGK LED PK KSGDA +V+++P KPMCVE+FS YPPLGRFAVRDM+ Sbjct: 371 KFAELKEKIDRRSGKKLEDNPKALKSGDAAIVEMVPGKPMCVESFSTYPPLGRFAVRDMR 430 Query: 1081 QTVAVGVI 1104 QTVAVGVI Sbjct: 431 QTVAVGVI 438
>sp|Q5R4R8|EF1A1_PONPY Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu) Length = 462 Score = 605 bits (1561), Expect = e-173 Identities = 288/368 (78%), Positives = 330/368 (89%) Frame = +1 Query: 1 ALWKFETTKFYVTVIDAPGHRDFIKNMITGTSQADCAVLVVAAGTGEFEAGISKNGQTRE 180 +LWKFET+K+YVT+IDAPGHRDFIKNMITGTSQADCAVL+VAAG GEFEAGISKNGQTRE Sbjct: 76 SLWKFETSKYYVTIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAGISKNGQTRE 135 Query: 181 HALLAYTLGVKQMIVGVNKMDSTEPPFSESRFNEIRKEVSSYLKKVGYQPDGVAFVPISG 360 HALLAYTLGVKQ+IVGVNKMDSTEPP+S+ R+ EI KEVS+Y+KK+GY PD VAFVPISG Sbjct: 136 HALLAYTLGVKQLIVGVNKMDSTEPPYSQKRYEEIVKEVSTYIKKIGYNPDTVAFVPISG 195 Query: 361 WNGDNMIEESSNMSWYKGWDITRKNAKKEETKITGKTLLDALDSLEPPSRPTNKPLRLPL 540 WNGDNM+E S+NM W+KGW +TRK+ +G TLL+ALD + PP+RPT+KPLRLPL Sbjct: 196 WNGDNMLEPSANMPWFKGWKVTRKDGNA-----SGTTLLEALDCILPPTRPTDKPLRLPL 250 Query: 541 QDVYKIGGIGTVPVGRVETGILKPGMVVTFAPQGLTTEVKSVEMHHEALTEALPGDNVGF 720 QDVYKIGGIGTVPVGRVETG+LKPG+VVTFAP +TTEVKSVEMHHEAL+EALPGDNVGF Sbjct: 251 QDVYKIGGIGTVPVGRVETGVLKPGVVVTFAPVNVTTEVKSVEMHHEALSEALPGDNVGF 310 Query: 721 NIKNVSVKEIRRGNVCSDSKSDPARQATDFIAQVIIMNHPGEIHSGYSPVLDCHTAHIAC 900 N+KNVSVK++RRGNV DSK+DP +A F AQVII+NHPG+I +GY+PVLDCHTAHIAC Sbjct: 311 NVKNVSVKDVRRGNVAGDSKNDPPMEAAGFTAQVIILNHPGQISAGYAPVLDCHTAHIAC 370 Query: 901 KFAELLKKIDRRSGKDLEDFPKQFKSGDAGLVKLIPSKPMCVETFSEYPPLGRFAVRDMK 1080 KFAEL +KIDRRSGK LED PK KSGDA +V ++P KPMCVE+FS+YPPLGRFAVRDM+ Sbjct: 371 KFAELKEKIDRRSGKKLEDGPKFLKSGDAAIVDMVPGKPMCVESFSDYPPLGRFAVRDMR 430 Query: 1081 QTVAVGVI 1104 QTVAVGVI Sbjct: 431 QTVAVGVI 438
>sp|Q71V39|EF1A2_RABIT Elongation factor 1-alpha 2 (EF-1-alpha-2) (Elongation factor 1 A-2) (eEF1A-2) (Statin S1) sp|Q05639|EF1A2_HUMAN Elongation factor 1-alpha 2 (EF-1-alpha-2) (Elongation factor 1 A-2) (eEF1A-2) (Statin S1) Length = 463 Score = 603 bits (1555), Expect = e-172 Identities = 287/368 (77%), Positives = 330/368 (89%) Frame = +1 Query: 1 ALWKFETTKFYVTVIDAPGHRDFIKNMITGTSQADCAVLVVAAGTGEFEAGISKNGQTRE 180 +LWKFETTK+Y+T+IDAPGHRDFIKNMITGTSQADCAVL+VAAG GEFEAGISKNGQTRE Sbjct: 76 SLWKFETTKYYITIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAGISKNGQTRE 135 Query: 181 HALLAYTLGVKQMIVGVNKMDSTEPPFSESRFNEIRKEVSSYLKKVGYQPDGVAFVPISG 360 HALLAYTLGVKQ+IVGVNKMDSTEP +SE R++EI KEVS+Y+KK+GY P V FVPISG Sbjct: 136 HALLAYTLGVKQLIVGVNKMDSTEPAYSEKRYDEIVKEVSAYIKKIGYNPATVPFVPISG 195 Query: 361 WNGDNMIEESSNMSWYKGWDITRKNAKKEETKITGKTLLDALDSLEPPSRPTNKPLRLPL 540 W+GDNM+E S NM W+KGW + RK E +G +LL+ALD++ PP+RPT+KPLRLPL Sbjct: 196 WHGDNMLEPSPNMPWFKGWKVERK-----EGNASGVSLLEALDTILPPTRPTDKPLRLPL 250 Query: 541 QDVYKIGGIGTVPVGRVETGILKPGMVVTFAPQGLTTEVKSVEMHHEALTEALPGDNVGF 720 QDVYKIGGIGTVPVGRVETGIL+PGMVVTFAP +TTEVKSVEMHHEAL+EALPGDNVGF Sbjct: 251 QDVYKIGGIGTVPVGRVETGILRPGMVVTFAPVNITTEVKSVEMHHEALSEALPGDNVGF 310 Query: 721 NIKNVSVKEIRRGNVCSDSKSDPARQATDFIAQVIIMNHPGEIHSGYSPVLDCHTAHIAC 900 N+KNVSVK+IRRGNVC DSKSDP ++A F +QVII+NHPG+I +GYSPV+DCHTAHIAC Sbjct: 311 NVKNVSVKDIRRGNVCGDSKSDPPQEAAQFTSQVIILNHPGQISAGYSPVIDCHTAHIAC 370 Query: 901 KFAELLKKIDRRSGKDLEDFPKQFKSGDAGLVKLIPSKPMCVETFSEYPPLGRFAVRDMK 1080 KFAEL +KIDRRSGK LED PK KSGDA +V+++P KPMCVE+FS+YPPLGRFAVRDM+ Sbjct: 371 KFAELKEKIDRRSGKKLEDNPKSLKSGDAAIVEMVPGKPMCVESFSQYPPLGRFAVRDMR 430 Query: 1081 QTVAVGVI 1104 QTVAVGVI Sbjct: 431 QTVAVGVI 438
>sp|P62631|EF1A2_MOUSE Elongation factor 1-alpha 2 (EF-1-alpha-2) (Elongation factor 1 A-2) (eEF1A-2) (Statin S1) sp|P62632|EF1A2_RAT Elongation factor 1-alpha 2 (EF-1-alpha-2) (Elongation factor 1 A-2) (eEF1A-2) (Statin S1) Length = 463 Score = 602 bits (1552), Expect = e-172 Identities = 286/368 (77%), Positives = 330/368 (89%) Frame = +1 Query: 1 ALWKFETTKFYVTVIDAPGHRDFIKNMITGTSQADCAVLVVAAGTGEFEAGISKNGQTRE 180 +LWKFETTK+Y+T+IDAPGHRDFIKNMITGTSQADCAVL+VAAG GEFEAGISKNGQTRE Sbjct: 76 SLWKFETTKYYITIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAGISKNGQTRE 135 Query: 181 HALLAYTLGVKQMIVGVNKMDSTEPPFSESRFNEIRKEVSSYLKKVGYQPDGVAFVPISG 360 HALLAYTLGVKQ+IVGVNKMDSTEP +SE R++EI KEVS+Y+KK+GY P V FVPISG Sbjct: 136 HALLAYTLGVKQLIVGVNKMDSTEPAYSEKRYDEIVKEVSAYIKKIGYNPATVPFVPISG 195 Query: 361 WNGDNMIEESSNMSWYKGWDITRKNAKKEETKITGKTLLDALDSLEPPSRPTNKPLRLPL 540 W+GDNM+E S NM W+KGW + RK E +G +LL+ALD++ PP+RPT+KPLRLPL Sbjct: 196 WHGDNMLEPSPNMPWFKGWKVERK-----EGNASGVSLLEALDTILPPTRPTDKPLRLPL 250 Query: 541 QDVYKIGGIGTVPVGRVETGILKPGMVVTFAPQGLTTEVKSVEMHHEALTEALPGDNVGF 720 QDVYKIGGIGTVPVGRVETGIL+PGMVVTFAP +TTEVKSVEMHHEAL+EALPGDNVGF Sbjct: 251 QDVYKIGGIGTVPVGRVETGILRPGMVVTFAPVNITTEVKSVEMHHEALSEALPGDNVGF 310 Query: 721 NIKNVSVKEIRRGNVCSDSKSDPARQATDFIAQVIIMNHPGEIHSGYSPVLDCHTAHIAC 900 N+KNVSVK+IRRGNVC DSK+DP ++A F +QVII+NHPG+I +GYSPV+DCHTAHIAC Sbjct: 311 NVKNVSVKDIRRGNVCGDSKADPPQEAAQFTSQVIILNHPGQISAGYSPVIDCHTAHIAC 370 Query: 901 KFAELLKKIDRRSGKDLEDFPKQFKSGDAGLVKLIPSKPMCVETFSEYPPLGRFAVRDMK 1080 KFAEL +KIDRRSGK LED PK KSGDA +V+++P KPMCVE+FS+YPPLGRFAVRDM+ Sbjct: 371 KFAELKEKIDRRSGKKLEDNPKSLKSGDAAIVEMVPGKPMCVESFSQYPPLGRFAVRDMR 430 Query: 1081 QTVAVGVI 1104 QTVAVGVI Sbjct: 431 QTVAVGVI 438
>sp|P17508|EF1A3_XENLA Elongation factor 1-alpha, oocyte form (EF-1-alpha-O1) (EF-1AO1) Length = 461 Score = 601 bits (1550), Expect = e-171 Identities = 288/368 (78%), Positives = 324/368 (88%) Frame = +1 Query: 1 ALWKFETTKFYVTVIDAPGHRDFIKNMITGTSQADCAVLVVAAGTGEFEAGISKNGQTRE 180 +LWKFET K+Y+T+IDAPGHRDFIKNMITGTSQADCAVL+VA G GEFEAGISKNGQTRE Sbjct: 76 SLWKFETGKYYITIIDAPGHRDFIKNMITGTSQADCAVLIVAGGVGEFEAGISKNGQTRE 135 Query: 181 HALLAYTLGVKQMIVGVNKMDSTEPPFSESRFNEIRKEVSSYLKKVGYQPDGVAFVPISG 360 HALLA+TLGVKQ+I+GVNKMDSTEPPFS+ RF EI KEVS+Y+KK+GY P V FVPISG Sbjct: 136 HALLAFTLGVKQLIIGVNKMDSTEPPFSQKRFEEITKEVSAYIKKIGYNPATVPFVPISG 195 Query: 361 WNGDNMIEESSNMSWYKGWDITRKNAKKEETKITGKTLLDALDSLEPPSRPTNKPLRLPL 540 W+GDNM+E S+NM W+KGW I RK E +G TLL+ALD + PP RPTNKPLRLPL Sbjct: 196 WHGDNMLEASTNMPWFKGWKIERK-----EGNASGITLLEALDCIIPPQRPTNKPLRLPL 250 Query: 541 QDVYKIGGIGTVPVGRVETGILKPGMVVTFAPQGLTTEVKSVEMHHEALTEALPGDNVGF 720 QDVYKIGGIGTVPVGRVETG+LKPGM+VTFAP +TTEVKSVEMHHEAL EALPGDNVGF Sbjct: 251 QDVYKIGGIGTVPVGRVETGVLKPGMIVTFAPSNVTTEVKSVEMHHEALVEALPGDNVGF 310 Query: 721 NIKNVSVKEIRRGNVCSDSKSDPARQATDFIAQVIIMNHPGEIHSGYSPVLDCHTAHIAC 900 N+KN+SVK+IRRGNV DSK+DP QA F AQVII+NHPG+I +GY+PVLDCHTAHIAC Sbjct: 311 NVKNISVKDIRRGNVAGDSKNDPPMQAGSFTAQVIILNHPGQISAGYAPVLDCHTAHIAC 370 Query: 901 KFAELLKKIDRRSGKDLEDFPKQFKSGDAGLVKLIPSKPMCVETFSEYPPLGRFAVRDMK 1080 KFAEL +KIDRRSGK LED PK KSGDA +V++IP KPMCVETFS+YPPLGRFAVRDM+ Sbjct: 371 KFAELKQKIDRRSGKKLEDDPKFLKSGDAAIVEMIPGKPMCVETFSDYPPLGRFAVRDMR 430 Query: 1081 QTVAVGVI 1104 QTVAVGVI Sbjct: 431 QTVAVGVI 438
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 140,899,053
Number of Sequences: 369166
Number of extensions: 3029854
Number of successful extensions: 11271
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9608
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10318
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 13626738475
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail