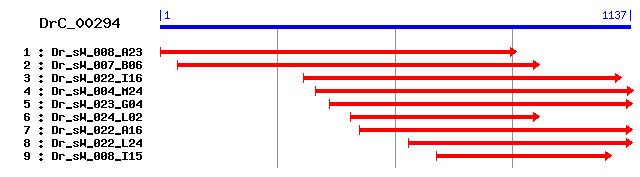
DrC_00294
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00294 (1136 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q05870|MYSP_SCHJA Paramyosin (Antigen Sj97) 385 e-107 sp|P06198|MYSP_SCHMA Paramyosin 383 e-106 sp|P35418|MYSP_TAESO Paramyosin (Antigen B) (AgB) 378 e-104 sp|P35417|MYSP_ECHGR Paramyosin 377 e-104 sp|Q8T305|MYSP_TAESA Paramyosin 376 e-104 sp|O96064|MYSP_MYTGA Paramyosin 309 8e-84 sp|P05661|MYSA_DROME Myosin heavy chain, muscle 215 2e-55 sp|P24733|MYS_AEQIR Myosin heavy chain, striated muscle 214 4e-55 sp|Q90339|MYSS_CYPCA Myosin heavy chain, fast skeletal muscle 211 3e-54 sp|P02566|MYO4_CAEEL Myosin-4 (Myosin heavy chain B) (MHC B... 209 9e-54
>sp|Q05870|MYSP_SCHJA Paramyosin (Antigen Sj97) Length = 866 Score = 385 bits (990), Expect = e-107 Identities = 199/285 (69%), Positives = 243/285 (85%) Frame = +3 Query: 66 SLLISERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKRR 245 +L I+E KR+QL E+++LR+ +E ER RK+AE E +E RVSELT+ VNTL NDKRR Sbjct: 587 NLQITEHKRIQLANEVEELRSAMENLERLRKHAETELEETQSRVSELTIQVNTLSNDKRR 646 Query: 246 LEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLEIE 425 LEGD+GV+Q D+DDA+NA+ A+EDRA +LNNE+ RLADELRQEQ+NYK A++LRK LEIE Sbjct: 647 LEGDIGVMQADMDDAINAKQAAEDRATRLNNEVLRLADELRQEQENYKHAEALRKQLEIE 706 Query: 426 IREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFKE 605 IREIT+KLEEAEA+ATREG+R +QKLQ R+RELE +F+ E RRCK+ALAQARKFERQ+KE Sbjct: 707 IREITVKLEEAEAFATREGRRMVQKLQARVRELEAEFDGESRRCKDALAQARKFERQYKE 766 Query: 606 LQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEHR 785 LQ QA+DDRRMV+ELQDLLDKTQ+KM+AYKRQLEE+E+VS + MNKYRKAQQQIE+AEHR Sbjct: 767 LQTQAEDDRRMVLELQDLLDKTQMKMKAYKRQLEEMEEVSQITMNKYRKAQQQIEEAEHR 826 Query: 786 ADMAERHLTVRRAGSVMPGSTYRAISVVRETSTVNTSRGARATSI 920 ADMAER +TVRR G PG RA+SV RE S V ++RG RATS+ Sbjct: 827 ADMAERTVTVRRVG---PGG--RAVSVARELS-VTSNRGMRATSM 865
Score = 75.9 bits (185), Expect = 2e-13
Identities = 65/250 (26%), Positives = 114/250 (45%), Gaps = 9/250 (3%)
Frame = +3
Query: 81 ERKRVQLQGELDDLRNQLEASE-------RARKNAENEAQELGGRVSELTLHVNTLINDK 239
E+ + +L E+ DL++++E+ R K+AE+ A EL RV ELT+ VNTL +
Sbjct: 331 EKLKTKLTLEIKDLQSEIESLSLENGELIRRAKSAESLASELQRRVDELTIEVNTLTSQN 390
Query: 240 RRLEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLE 419
+LE + ++ ++D + A E ++N+++ L LR ++LR LE
Sbjct: 391 NQLESENMRLKSLVNDLTDKNNALERENRQMNDQVKELKSSLRDANRRLTDLEALRSQLE 450
Query: 420 IEIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARK-FERQ 596
E + L +AE A R+ + Q Q L L+++ E R E L RK R
Sbjct: 451 AERDNLASALHDAEE-ALRDMDQKYQASQAALNHLKSEMEQRLRERDEELESLRKSTTRT 509
Query: 597 FKELQQQADD-DRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIED 773
+EL + + + EL L + + + + QL+ + M + + Q+++D
Sbjct: 510 IEELTVTITEMEVKYKSELSRLKKRYESSIADLEIQLDATNKANANLMKENKNLAQRVKD 569
Query: 774 AEHRADMAER 803
E D R
Sbjct: 570 LETFLDDERR 579
>sp|P06198|MYSP_SCHMA Paramyosin Length = 866 Score = 383 bits (983), Expect = e-106 Identities = 200/285 (70%), Positives = 242/285 (84%) Frame = +3 Query: 66 SLLISERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKRR 245 +L I+E KR+QL E++++R+ LE ER RK+AE E +E RVSELT+ VNTL NDKRR Sbjct: 587 NLQITEHKRLQLANEIEEIRSTLENLERLRKHAETELEEAQSRVSELTIQVNTLTNDKRR 646 Query: 246 LEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLEIE 425 LEGD+GV+Q D+DDA+NA+ ASEDRA +LNNE+ RLADELRQEQ NYK A++LRK LEIE Sbjct: 647 LEGDIGVMQADMDDAINAKQASEDRAIRLNNEVLRLADELRQEQGNYKHAEALRKQLEIE 706 Query: 426 IREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFKE 605 IREIT+KLEEAEA ATREG+R +QKLQ R+RELE++F+ E RRCK+ALAQARKFERQ+KE Sbjct: 707 IREITVKLEEAEASATREGRRMVQKLQARVRELESEFDGESRRCKDALAQARKFERQYKE 766 Query: 606 LQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEHR 785 LQ QA+DDRRMV+ELQDLLDKTQ+KM+AYKRQLEE+E+VS + MNKYRKAQQQIE+AEHR Sbjct: 767 LQTQAEDDRRMVLELQDLLDKTQMKMKAYKRQLEEMEEVSQITMNKYRKAQQQIEEAEHR 826 Query: 786 ADMAERHLTVRRAGSVMPGSTYRAISVVRETSTVNTSRGARATSI 920 ADMAER +TVRR G PG RA+SV RE S V ++RG RATS+ Sbjct: 827 ADMAERTVTVRRVG---PGG--RAVSVARELS-VTSNRGMRATSM 865
Score = 69.7 bits (169), Expect = 1e-11
Identities = 62/250 (24%), Positives = 111/250 (44%), Gaps = 9/250 (3%)
Frame = +3
Query: 81 ERKRVQLQGELDDLRNQLEASE-------RARKNAENEAQELGGRVSELTLHVNTLINDK 239
E+ + +L E+ DL++++E+ R K AE+ A +L RV ELT+ VNTL +
Sbjct: 331 EKLKTKLTLEIKDLQSEIESLSLENSELIRRAKAAESLASDLQRRVDELTIEVNTLTSQN 390
Query: 240 RRLEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLE 419
+LE + ++ ++D + E ++N+++ L LR ++LR LE
Sbjct: 391 SQLESENLRLKSLVNDLTDKNNLLERENRQMNDQVKELKSSLRDANRRLTDLEALRSQLE 450
Query: 420 IEIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARK-FERQ 596
E + L +AE A + + Q Q L L+++ E R E L RK R
Sbjct: 451 AERDNLASALHDAEE-ALHDMDQKYQASQAALNHLKSEMEQRLRERDEELESLRKSTTRT 509
Query: 597 FKELQQQADD-DRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIED 773
+EL + + + EL L + + + + QL+ + M + + Q+++D
Sbjct: 510 IEELTVTITEMEVKYKSELSRLKKRYESNIADLEIQLDTANKANANLMKENKNLSQRVKD 569
Query: 774 AEHRADMAER 803
E D R
Sbjct: 570 LETFLDEERR 579
>sp|P35418|MYSP_TAESO Paramyosin (Antigen B) (AgB) Length = 863 Score = 378 bits (971), Expect = e-104 Identities = 195/285 (68%), Positives = 244/285 (85%) Frame = +3 Query: 66 SLLISERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKRR 245 +L +SERKR+ L E++++R+QLE S+RARKNAE+E + GR+SELTL VNTL NDKRR Sbjct: 583 NLQVSERKRIALASEVEEIRSQLELSDRARKNAESELNDANGRISELTLSVNTLTNDKRR 642 Query: 246 LEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLEIE 425 LEGD+GV+QGDLD+AVNAR A+EDRA++LN E+ RLADELRQEQ+NYK+A++LRK LEIE Sbjct: 643 LEGDIGVMQGDLDEAVNARKAAEDRADRLNAEVLRLADELRQEQENYKRAETLRKQLEIE 702 Query: 426 IREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFKE 605 IREIT+KLEEAEA+ATREG+R +QKLQ R+RELE + + E RR KEA A ARK+ERQFKE Sbjct: 703 IREITVKLEEAEAFATREGRRMVQKLQNRVRELEAELDGEIRRAKEAFANARKYERQFKE 762 Query: 606 LQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEHR 785 LQ Q++DD+RM++ELQDLLDKTQ+KM+AYKRQLEE E+VS + M+KYRKAQQQIE+AEHR Sbjct: 763 LQTQSEDDKRMILELQDLLDKTQIKMKAYKRQLEEQEEVSQLTMSKYRKAQQQIEEAEHR 822 Query: 786 ADMAERHLTVRRAGSVMPGSTYRAISVVRETSTVNTSRGARATSI 920 ADMAER +T++R + G RA+SVVRE ++V SRG RATSI Sbjct: 823 ADMAERTITIKR---TIGGPGSRAVSVVREINSV--SRGNRATSI 862
Score = 72.8 bits (177), Expect = 2e-12
Identities = 75/266 (28%), Positives = 121/266 (45%), Gaps = 21/266 (7%)
Frame = +3
Query: 81 ERKRVQLQGELDDLRNQLEA--------SERARKNAENEAQELGGRVSELTLHVNTLIND 236
E+ +V+L E+ DL+ + EA + RA K AEN A EL R+ E+T+ +NTL +
Sbjct: 327 EKTKVKLTLEIKDLQAENEALAAENGELTHRA-KQAENLANELQRRIDEMTVEINTLNSA 385
Query: 237 KRRLEGDLGVIQGDLDDAVNARLASEDRANK-LNNELNRLADELRQEQDNYKKADSLRKT 413
LE D ++G + D + R+A+ DR N+ L ++L LR ++LR
Sbjct: 386 NSALEADNMRLKGQVGDLTD-RIANLDRENRQLGDQLKETKSALRDANRRLTDLEALRSQ 444
Query: 414 LEIEIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARK-FE 590
LE E + L +AE A +E + Q L L+++ E R E L RK
Sbjct: 445 LEAERDNLASALHDAEE-ALKEMEAKYVASQNALNHLKSEMEQRLREKDEELENLRKSTT 503
Query: 591 RQFKELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLE---EIEDVSTVAMNKYRKA-- 755
R +EL + M V + + + + K A +LE ++ + + V +N+ K
Sbjct: 504 RTIEELTTTISE---MEVRFKSDMSRLKKKYEATISELEVQLDVANKANVNLNRENKTLA 560
Query: 756 ------QQQIEDAEHRADMAERHLTV 815
Q +ED + AE +L V
Sbjct: 561 QRVQELQAALEDERRAREAAESNLQV 586
Score = 46.2 bits (108), Expect = 2e-04
Identities = 45/246 (18%), Positives = 96/246 (39%), Gaps = 14/246 (5%)
Frame = +3
Query: 84 RKRVQLQGELDDLRNQLEASERARKNAENEA-------QELGGRVSELTLHVNTLINDKR 242
+ + L+ +LDDL+ ++ R R + + + L R E L N
Sbjct: 215 KTKAALESQLDDLKRAMDEDARNRLSLQTQLSSLQMDYDNLQARYEEEAEAAGNLRNQVA 274
Query: 243 RLEGDLGVIQGDLDDAVNARLAS-EDRANKLNNELNRLADELRQEQDNYKKADSLRKTLE 419
+ D+ ++ L+ + A+ E+ KL + L D E+ + + L
Sbjct: 275 KFNADMAALKTRLERELMAKTEEFEELKRKLTVRITELEDMAEHERTRANNLEKTKVKLT 334
Query: 420 IEIREITIKLEEAEA------YATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQAR 581
+EI+++ + E A + ++ + +LQ R+ E+ + A
Sbjct: 335 LEIKDLQAENEALAAENGELTHRAKQAENLANELQRRIDEMTVEINTLNSANSALEADNM 394
Query: 582 KFERQFKELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQ 761
+ + Q +L + + R +L D L +T+ +R R+L ++E + + +
Sbjct: 395 RLKGQVGDLTDRIANLDRENRQLGDQLKETKSALRDANRRLTDLEALRSQLEAERDNLAS 454
Query: 762 QIEDAE 779
+ DAE
Sbjct: 455 ALHDAE 460
>sp|P35417|MYSP_ECHGR Paramyosin Length = 863 Score = 377 bits (969), Expect = e-104 Identities = 194/285 (68%), Positives = 244/285 (85%) Frame = +3 Query: 66 SLLISERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKRR 245 +L +SERKR+ L E++++R+QLE S+RARKNAE+E + GR+SELT+ VNTL NDKRR Sbjct: 583 NLQVSERKRIALTSEVEEIRSQLELSDRARKNAESELNDANGRISELTMSVNTLTNDKRR 642 Query: 246 LEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLEIE 425 LEGD+GV+QGDLD+AVNAR A+EDRA++LN E+ RLADELRQEQ+NYK+A++LRK LEIE Sbjct: 643 LEGDIGVMQGDLDEAVNARKAAEDRADRLNAEVLRLADELRQEQENYKRAETLRKQLEIE 702 Query: 426 IREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFKE 605 IREIT+KLEEAEA+ATREG+R +QKLQ R+RELE + + E RR KEA A ARK+ERQFKE Sbjct: 703 IREITVKLEEAEAFATREGRRMVQKLQNRVRELEAELDGEIRRAKEAFASARKYERQFKE 762 Query: 606 LQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEHR 785 LQ Q++DD+RM++ELQDLLDKTQ+KM+AYKRQLEE E+VS + M+KYRKAQQQIE+AEHR Sbjct: 763 LQTQSEDDKRMILELQDLLDKTQIKMKAYKRQLEEQEEVSQLTMSKYRKAQQQIEEAEHR 822 Query: 786 ADMAERHLTVRRAGSVMPGSTYRAISVVRETSTVNTSRGARATSI 920 ADMAER +T++R + G RA+SVVRE ++V SRG RATSI Sbjct: 823 ADMAERTITIKR---TIGGPGSRAVSVVREINSV--SRGNRATSI 862
Score = 74.3 bits (181), Expect = 6e-13
Identities = 76/266 (28%), Positives = 123/266 (46%), Gaps = 21/266 (7%)
Frame = +3
Query: 81 ERKRVQLQGELDDLRNQLEA--------SERARKNAENEAQELGGRVSELTLHVNTLIND 236
E+ +V+L E+ DL+ + EA + RA K AEN A EL RV E+T+ +NTL +
Sbjct: 327 EKTKVKLTIEIKDLQAENEALAAENGELTHRA-KQAENLANELQRRVDEMTVEINTLNSA 385
Query: 237 KRRLEGDLGVIQGDLDDAVNARLASEDRANK-LNNELNRLADELRQEQDNYKKADSLRKT 413
LEGD ++G + D + R+A+ DR N+ L ++L LR ++LR
Sbjct: 386 NNALEGDNMRLKGQVGDLTD-RIANLDRENRQLGDQLKETKSALRDANRRLTDLEALRSQ 444
Query: 414 LEIEIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARK-FE 590
LE + + L +AE A +E + Q L L+++ E R E L RK
Sbjct: 445 LEADGDNLASALHDAEE-ALKELEVKYVASQNALNHLKSEMEQRLREKDEELENLRKSTT 503
Query: 591 RQFKELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLE---EIEDVSTVAMNKYRKA-- 755
R +EL + M V + + + + K A +LE ++ + + ++N+ KA
Sbjct: 504 RTIEELTTTISE---MEVRFKSDMSRLKKKYEATISELEVQLDVANKANASLNRENKALA 560
Query: 756 ------QQQIEDAEHRADMAERHLTV 815
Q +ED + AE +L V
Sbjct: 561 QRVQELQTALEDERRAREAAESNLQV 586
Score = 49.3 bits (116), Expect = 2e-05
Identities = 53/259 (20%), Positives = 102/259 (39%), Gaps = 15/259 (5%)
Frame = +3
Query: 99 LQGELDDLRNQLEASERARKNAENEA-------QELGGRVSELTLHVNTLINDKRRLEGD 257
L+G+LDDL+ ++ R R N + + L R E L N + D
Sbjct: 220 LEGQLDDLKRAMDEDARNRLNLQTQLSSLQMDYDNLQARYEEEAEAAGNLRNQVAKFNAD 279
Query: 258 LGVIQGDLDDAVNARLAS-EDRANKLNNELNRLADELRQEQDNYKKADSLRKTLEIEIRE 434
+ ++ L+ + A+ E+ KL + L D E+ + + L IEI++
Sbjct: 280 MAALKTRLERELMAKTEEFEELKRKLTVRITELEDIAEHERTRANNLEKTKVKLTIEIKD 339
Query: 435 ITIKLEEAEA------YATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQ 596
+ + E A + ++ + +LQ R+ E+ + + + Q
Sbjct: 340 LQAENEALAAENGELTHRAKQAENLANELQRRVDEMTVEINTLNSANNALEGDNMRLKGQ 399
Query: 597 FKELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQ-QQIED 773
+L + + R +L D L +T+ +R R+L ++E A+ +A +
Sbjct: 400 VGDLTDRIANLDRENRQLGDQLKETKSALRDANRRLTDLE-----ALRSQLEADGDNLAS 454
Query: 774 AEHRADMAERHLTVRRAGS 830
A H A+ A + L V+ S
Sbjct: 455 ALHDAEEALKELEVKYVAS 473
>sp|Q8T305|MYSP_TAESA Paramyosin Length = 863 Score = 376 bits (965), Expect = e-104 Identities = 194/285 (68%), Positives = 243/285 (85%) Frame = +3 Query: 66 SLLISERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKRR 245 +L +SERKR+ L E++++R+QLE S+RARKNAE+E + GR+SELTL VNTL NDKRR Sbjct: 583 NLQVSERKRIALASEVEEIRSQLELSDRARKNAESELNDANGRISELTLSVNTLTNDKRR 642 Query: 246 LEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLEIE 425 LEGD+GV+QGDLD+AVNAR A+EDRA++LN E+ RLADELRQEQ+NY +A++LRK LEIE Sbjct: 643 LEGDIGVMQGDLDEAVNARKAAEDRADRLNAEVLRLADELRQEQENYXRAETLRKQLEIE 702 Query: 426 IREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFKE 605 IREIT+KLEEAEA+ATREG+R +QKLQ R+RELE + + E RR KEA A ARK+ERQFKE Sbjct: 703 IREITVKLEEAEAFATREGRRMVQKLQNRVRELEAELDGEIRRAKEAFANARKYERQFKE 762 Query: 606 LQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEHR 785 LQ Q++DD+RM++ELQDLLDKTQ+KM+AYKRQLEE E+VS + M+KYRKAQQQIE+AEHR Sbjct: 763 LQTQSEDDKRMILELQDLLDKTQIKMKAYKRQLEEQEEVSQLTMSKYRKAQQQIEEAEHR 822 Query: 786 ADMAERHLTVRRAGSVMPGSTYRAISVVRETSTVNTSRGARATSI 920 ADMAER +T++R + G RA+SVVRE ++V SRG RATSI Sbjct: 823 ADMAERTITIKR---TIGGPGSRAVSVVREINSV--SRGNRATSI 862
Score = 72.4 bits (176), Expect = 2e-12
Identities = 76/265 (28%), Positives = 119/265 (44%), Gaps = 20/265 (7%)
Frame = +3
Query: 81 ERKRVQLQGELDDLR--NQLEASERAR-----KNAENEAQELGGRVSELTLHVNTLINDK 239
E+ +V+L E+ DL+ N+ A+E K AEN A EL R+ E+T+ +NTL +
Sbjct: 327 EKTKVKLTLEIKDLQAENEALAAENGELTHRVKQAENLANELQRRIDEMTVEINTLNSAN 386
Query: 240 RRLEGDLGVIQGDLDDAVNARLASEDRANK-LNNELNRLADELRQEQDNYKKADSLRKTL 416
LE D ++G + D + R+A+ DR N+ L ++L LR ++LR L
Sbjct: 387 SALEADNMRLKGQVGDLTD-RIANLDRENRQLGDQLKETKSALRDANRRLTDLEALRSQL 445
Query: 417 EIEIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARK-FER 593
E E + L +AE A +E + Q L L+++ E R E L RK R
Sbjct: 446 EAERDNLASALHDAEE-ALKEMEAKYVASQNALNHLKSEMEQRLREKDEELENLRKSTTR 504
Query: 594 QFKELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVA---MNKYRKA--- 755
+EL + M V + + + + K A +LE DV+ A +N+ K
Sbjct: 505 TIEELTTTISE---MEVRFKSDMSRLKKKYEATISELEVQLDVANKANANLNRENKTLAQ 561
Query: 756 -----QQQIEDAEHRADMAERHLTV 815
Q +ED + AE +L V
Sbjct: 562 RVQELQAALEDERRAREAAESNLQV 586
Score = 48.1 bits (113), Expect = 5e-05
Identities = 46/246 (18%), Positives = 96/246 (39%), Gaps = 14/246 (5%)
Frame = +3
Query: 84 RKRVQLQGELDDLRNQLEASERARKNAENEA-------QELGGRVSELTLHVNTLINDKR 242
+ + L+ +LDDL+ ++ R R N + + L R E L N
Sbjct: 215 KTKAALESQLDDLKRAMDEDARNRLNLQTQLSSLQMDYDNLQARYEEEAEAAGNLRNQVA 274
Query: 243 RLEGDLGVIQGDLDDAVNARLAS-EDRANKLNNELNRLADELRQEQDNYKKADSLRKTLE 419
+ D+ ++ L+ + A+ E+ KL + L D E+ + + L
Sbjct: 275 KFNADMAALKTRLERELMAKTEEFEELKRKLTVRITELEDMAEHERTRANNLEKTKVKLT 334
Query: 420 IEIREITIKLEEAEA------YATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQAR 581
+EI+++ + E A + ++ + +LQ R+ E+ + A
Sbjct: 335 LEIKDLQAENEALAAENGELTHRVKQAENLANELQRRIDEMTVEINTLNSANSALEADNM 394
Query: 582 KFERQFKELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQ 761
+ + Q +L + + R +L D L +T+ +R R+L ++E + + +
Sbjct: 395 RLKGQVGDLTDRIANLDRENRQLGDQLKETKSALRDANRRLTDLEALRSQLEAERDNLAS 454
Query: 762 QIEDAE 779
+ DAE
Sbjct: 455 ALHDAE 460
>sp|O96064|MYSP_MYTGA Paramyosin Length = 864 Score = 309 bits (792), Expect = 8e-84 Identities = 162/278 (58%), Positives = 210/278 (75%), Gaps = 1/278 (0%) Frame = +3 Query: 57 TNESLLISERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLIND 236 T L +SERKR+ +Q EL+D R+ LE +ERARKNAENE E+ R++E+ L V L ND Sbjct: 593 TRNQLSVSERKRITIQQELEDARSLLEHAERARKNAENELGEVTVRLTEVQLQVTALTND 652 Query: 237 KRRLEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTL 416 KRR+E D+ +Q DLDDA+N + A+E+RA++L E+NRLADELRQEQDNYK A+SLRK L Sbjct: 653 KRRMEADIAAMQSDLDDALNGQRAAEERADRLQAEVNRLADELRQEQDNYKNAESLRKQL 712 Query: 417 EIEIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQ 596 EIEIREIT++LEEAEA+A REGKR I KLQ R+R+LE + EA+QRR +EA A ARKFERQ Sbjct: 713 EIEIREITVRLEEAEAFAQREGKRQIAKLQARIRDLENELEADQRRLREAAASARKFERQ 772 Query: 597 FKELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDA 776 +KE Q +D+DRR V EL + D+ +K + YKR +EE EDV+++ MNKYRKAQ I++A Sbjct: 773 WKETVQASDEDRRQVAELTSITDQLTMKCKTYKRMIEEAEDVASITMNKYRKAQSLIDEA 832 Query: 777 EHRADMAERHL-TVRRAGSVMPGSTYRAISVVRETSTV 887 E RADMAE++L TVRR+ R++SV RE + V Sbjct: 833 EQRADMAEKNLQTVRRS---------RSMSVSREVTRV 861
Score = 62.8 bits (151), Expect = 2e-09
Identities = 50/224 (22%), Positives = 97/224 (43%), Gaps = 1/224 (0%)
Frame = +3
Query: 144 ERARKNAENEAQELGGRVSELTLHVNTLINDKRRLEGDLGVIQGDLDDAVNARLASEDRA 323
ER +N +E + +N L K +LE + + L D A +E +
Sbjct: 424 ERENAGLQNALREANNELKAANRTINELTALKAQLEAERDNLASALRDTEEALRDAEAKL 483
Query: 324 NKLNNELNRLADELRQE-QDNYKKADSLRKTLEIEIREITIKLEEAEAYATREGKRAIQK 500
LN+L E+ Q ++ ++ DS+RK+ I E+ L E E E R +K
Sbjct: 484 AAAQAALNQLRSEMEQRLREKDEEIDSIRKSSARAIDELQRTLVEVETRYKTEISRIKKK 543
Query: 501 LQVRLRELETDFEAEQRRCKEALAQARKFERQFKELQQQADDDRRMVVELQDLLDKTQLK 680
+ +RELE + R E L Q + + + +EL+ Q ++ R + + ++ L ++ K
Sbjct: 544 YETDIRELEGALDNANRANAEYLKQIKSLQNRNRELELQLEEATRQLDDTRNQLSVSERK 603
Query: 681 MRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEHRADMAERHLT 812
+++LE+ + A + A+ ++ + R + +T
Sbjct: 604 RITIQQELEDARSLLEHAERARKNAENELGEVTVRLTEVQLQVT 647
Score = 56.6 bits (135), Expect = 1e-07
Identities = 63/262 (24%), Positives = 108/262 (41%), Gaps = 23/262 (8%)
Frame = +3
Query: 81 ERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKRRLEGDL 260
ER+ LQ L + N+L+A+ R ++ELT L ++ L L
Sbjct: 424 ERENAGLQNALREANNELKAANRT--------------INELTALKAQLEAERDNLASAL 469
Query: 261 GVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKA-----DSLRKTL--- 416
+ L DA A++ N+L +E+ + E +E D+ +K+ D L++TL
Sbjct: 470 RDTEEALRDAEAKLAAAQAALNQLRSEMEQRLREKDEEIDSIRKSSARAIDELQRTLVEV 529
Query: 417 ---------------EIEIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQR 551
E +IRE+ L+ A A E + I+ LQ R RELE E R
Sbjct: 530 ETRYKTEISRIKKKYETDIRELEGALDNANR-ANAEYLKQIKSLQNRNRELELQLEEATR 588
Query: 552 RCKEALAQARKFERQFKELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTV 731
+ + Q ER+ +QQ+ +D R ++ + + ++ +L E++ T
Sbjct: 589 QLDDTRNQLSVSERKRITIQQELEDARSLLEHAERARKNAENELGEVTVRLTEVQLQVTA 648
Query: 732 AMNKYRKAQQQIEDAEHRADMA 797
N R+ + I + D A
Sbjct: 649 LTNDKRRMEADIAAMQSDLDDA 670
Score = 46.6 bits (109), Expect = 1e-04
Identities = 51/281 (18%), Positives = 120/281 (42%), Gaps = 4/281 (1%)
Frame = +3
Query: 81 ERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKRRLEGDL 260
E +L+ +DDL QL S ++ E +L +V EL L K +L+
Sbjct: 178 EGSNSRLKAAVDDLTRQLNDSNLSKARLTQENFDLQHQVQELDGANAGLAKAKAQLQILC 237
Query: 261 GVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLEIEIREIT 440
++ +LDD R + + + ++ + L +E +N + +L +
Sbjct: 238 DDLKRNLDDESRQRQNLQVQLAAIQSDYDNLNARYEEESENASSLRAQLSSLSATYAALK 297
Query: 441 IKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFKELQQQA 620
K ++ E + ++L ++++ELE E + RC K + +E+ +
Sbjct: 298 TKYDKELMAKQEELEEIRRRLSIKIQELEDTCEQLRTRCNTLEKTKNKLTVEIREITIEL 357
Query: 621 DDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEHR---AD 791
++ + +V +L + + + A +++ +++ + N+ KA + E A + A+
Sbjct: 358 ENTQIIVQDLTKRNRQLENENAALQKRCDDLSAENGQLRNE--KANLEAECARLKVANAE 415
Query: 792 MAERHLTVRRAGSVMPGSTYRAISVVRETS-TVNTSRGARA 911
+AE++ + R + + + A + ++ + T+N +A
Sbjct: 416 LAEKNANLERENAGLQNALREANNELKAANRTINELTALKA 456
Score = 35.8 bits (81), Expect = 0.23
Identities = 35/188 (18%), Positives = 79/188 (42%), Gaps = 3/188 (1%)
Frame = +3
Query: 231 NDKRRLEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRK 410
+ + RLE + ++ LD RL E ++ +L +L+++AD L + +
Sbjct: 31 SSQNRLESRIRELEDSLDSEREMRLRYEKQSAELTFQLDQMADRLEDAMGTSTTVSEVSR 90
Query: 411 TLEIEIREI--TIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARK 584
E E+ ++ ++L A+ AT + R ++ Q L +L E + A + +
Sbjct: 91 KREAEVNKVRKDLELSCAQFEATEQNMR--RRHQEALNDLTDQLEHMGKSKSRAEKEKNQ 148
Query: 585 FERQFKELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEI-EDVSTVAMNKYRKAQQ 761
+ LQ D ++ + +D + K ++++ ++ ++K R Q+
Sbjct: 149 LIIEIDSLQAMNDGLQKSKMSADSKIDALEGSNSRLKAAVDDLTRQLNDSNLSKARLTQE 208
Query: 762 QIEDAEHR 785
D +H+
Sbjct: 209 NF-DLQHQ 215
>sp|P05661|MYSA_DROME Myosin heavy chain, muscle Length = 1962 Score = 215 bits (547), Expect = 2e-55 Identities = 123/287 (42%), Positives = 185/287 (64%) Frame = +3 Query: 63 ESLLISERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKR 242 E L ISER+ LQ EL++ R LE ++R R+ AE E + +++E++ ++ KR Sbjct: 1668 EQLGISERRANALQNELEESRTLLEQADRGRRQAEQELADAHEQLNEVSAQNASISAAKR 1727 Query: 243 RLEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLEI 422 +LE +L + DLD+ +N SE++A K + RLADELR EQD+ + + LRK LE Sbjct: 1728 KLESELQTLHSDLDELLNEAKNSEEKAKKAMVDAARLADELRAEQDHAQTQEKLRKALEQ 1787 Query: 423 EIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFK 602 +I+E+ ++L+EAEA A + GK+AIQKL+ R+RELE + + EQRR +A RK ER+ K Sbjct: 1788 QIKELQVRLDEAEANALKGGKKAIQKLEQRVRELENELDGEQRRHADAQKNLRKSERRVK 1847 Query: 603 ELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEH 782 EL Q+++DR+ +QDL+DK Q K++ YKRQ+EE E+++ + + K+RKAQQ++E+AE Sbjct: 1848 ELSFQSEEDRKNHERMQDLVDKLQQKIKTYKRQIEEAEEIAALNLAKFRKAQQELEEAEE 1907 Query: 783 RADMAERHLTVRRAGSVMPGSTYRAISVVRETSTVNTSRGARATSIR 923 RAD+AE+ ++ RA RA SV R S RATS+R Sbjct: 1908 RADLAEQAISKFRAKG-------RAGSVGR-----GASPAPRATSVR 1942
Score = 85.1 bits (209), Expect = 3e-16
Identities = 56/231 (24%), Positives = 122/231 (52%), Gaps = 1/231 (0%)
Frame = +3
Query: 96 QLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKRRLEGDLGVIQG 275
+L+G ++ + QLEA R KN +E ++L ++ E +++ + ++RLE + +Q
Sbjct: 1482 RLKGAYEEGQEQLEAVRRENKNLADEVKDLLDQIGEGGRNIHEIEKARKRLEAEKDELQA 1541
Query: 276 DLDDAVNARLASEDRANKLNNELNRLADEL-RQEQDNYKKADSLRKTLEIEIREITIKLE 452
L++A A E++ + EL+++ E+ R+ Q+ ++ ++ RK + + + LE
Sbjct: 1542 ALEEAEAALEQEENKVLRAQLELSQVRQEIDRRIQEKEEEFENTRKNHQRALDSMQASLE 1601
Query: 453 EAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFKELQQQADDDR 632
AEA E R +KL+ + ELE + + EA +++++Q K++Q ++++
Sbjct: 1602 -AEAKGKAEALRMKKKLEADINELEIALDHANKANAEAQKNIKRYQQQLKDIQTALEEEQ 1660
Query: 633 RMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEHR 785
R + ++ L ++ + A + +LEE + A R+A+Q++ DA +
Sbjct: 1661 RARDDAREQLGISERRANALQNELEESRTLLEQADRGRRQAEQELADAHEQ 1711
Score = 64.3 bits (155), Expect = 6e-10
Identities = 55/241 (22%), Positives = 109/241 (45%), Gaps = 5/241 (2%)
Frame = +3
Query: 96 QLQGELDDLRNQLEASERARKNAENEA---QELGGRVSELTLHVNTLINDKRRLEGDLGV 266
Q++ D++ +Q E + K + + Q+ G + +N L K +LE L
Sbjct: 973 QIRNLNDEIAHQDELINKLNKEKKMQGETNQKTGEELQAAEDKINHLNKVKAKLEQTLDE 1032
Query: 267 IQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLEIEIREITIK 446
++ L+ R E K+ +L + + + N K+ + + + E+ IT K
Sbjct: 1033 LEDSLEREKKVRGDVEKSKRKVEGDLKLTQEAVADLERNKKELEQTIQRKDKELSSITAK 1092
Query: 447 LEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFKELQQQADD 626
LE+ + + +R I++LQ R+ ELE + EAE++ +A Q R+ +EL ++ ++
Sbjct: 1093 LEDEQVVVLKH-QRQIKELQARIEELEEEVEAERQARAKAEKQRADLARELEELGERLEE 1151
Query: 627 DRRMVVELQDLLDKTQLKMRAYKRQLEE--IEDVSTVAMNKYRKAQQQIEDAEHRADMAE 800
+L K + ++ +R LEE I+ ST+A N +K + + + D
Sbjct: 1152 AGGATSAQIELNKKREAELSKLRRDLEEANIQHESTLA-NLRKKHNDAVAEMAEQVDQLN 1210
Query: 801 R 803
+
Sbjct: 1211 K 1211
Score = 63.9 bits (154), Expect = 8e-10
Identities = 66/290 (22%), Positives = 126/290 (43%), Gaps = 36/290 (12%)
Frame = +3
Query: 60 NESLLISERKRVQLQGELDDLRNQLE-------ASERARKNAENEAQELGGRVSELTLHV 218
N+ + E+ + +L E++DL+ +++ A+E+ +K + E +V +L +
Sbjct: 1407 NQKCIGLEKTKQRLSTEVEDLQLEVDRANAIANAAEKKQKAFDKIIGEWKLKVDDLAAEL 1466
Query: 219 NTLINDKRRLEGDLGVIQGDLDDAVNARLASEDRANK-LNNELNRLADELRQEQDNYKKA 395
+ + R +L ++G ++ +L + R NK L +E+ L D++ + N +
Sbjct: 1467 DASQKECRNYSTELFRLKGAYEEG-QEQLEAVRRENKNLADEVKDLLDQIGEGGRNIHEI 1525
Query: 396 DSLRKTLEIEIREITIKLEEAEAYATREGKRAI----------QKLQVRLRELETDF--- 536
+ RK LE E E+ LEEAEA +E + + Q++ R++E E +F
Sbjct: 1526 EKARKRLEAEKDELQAALEEAEAALEQEENKVLRAQLELSQVRQEIDRRIQEKEEEFENT 1585
Query: 537 ---------------EAEQRRCKEALAQARKFERQFKELQQQADDDRRMVVELQDLLDKT 671
EAE + EAL +K E EL+ D + E
Sbjct: 1586 RKNHQRALDSMQASLEAEAKGKAEALRMKKKLEADINELEIALDHANKANAE-------A 1638
Query: 672 QLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEHRADMAERHLTVRR 821
Q ++ Y++QL++I+ A++Q+ +E RA+ + L R
Sbjct: 1639 QKNIKRYQQQLKDIQTALEEEQRARDDAREQLGISERRANALQNELEESR 1688
Score = 44.7 bits (104), Expect = 5e-04
Identities = 48/220 (21%), Positives = 92/220 (41%), Gaps = 4/220 (1%)
Frame = +3
Query: 144 ERARKNAENEAQELGGRVSELTLHVNTLINDKRRLEGDLGVIQGDLDDAVNARLASEDRA 323
E+A+K E A E+ R EL L+ +K L L +G L D +
Sbjct: 853 EKAKKAEELHAAEVKVR-KELEALNAKLLAEKTALLDSLSGEKGALQDYQERNAKLTAQK 911
Query: 324 NKLNNELNRLADELRQEQDNYKKADSLRKTLEIEIREITIKLEEAEAYATREGKRAIQKL 503
N L N+L + + L QE+D + +K + EI + K+ I+ L
Sbjct: 912 NDLENQLRDIQERLTQEEDARNQLFQQKKKADQEISGL---------------KKDIEDL 956
Query: 504 QVRLRELETDFEAEQRRCK----EALAQARKFERQFKELQQQADDDRRMVVELQDLLDKT 671
++ +++ E D + + + E Q + KE + Q + +++ ELQ DK
Sbjct: 957 ELNVQKAEQDKATKDHQIRNLNDEIAHQDELINKLNKEKKMQGETNQKTGEELQAAEDKI 1016
Query: 672 QLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEHRAD 791
+ K +LE+ D ++ + +K + +E ++ + +
Sbjct: 1017 N-HLNKVKAKLEQTLDELEDSLEREKKVRGDVEKSKRKVE 1055
Score = 44.7 bits (104), Expect = 5e-04
Identities = 52/248 (20%), Positives = 100/248 (40%), Gaps = 23/248 (9%)
Frame = +3
Query: 96 QLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKRRLEGDLGVIQG 275
++Q +LD+ L + ++K E +L ++ E V+ L K L L +
Sbjct: 1257 EVQSKLDETNRTLNDFDASKKKLSIENSDLLRQLEEAESQVSQLSKIKISLTTQLEDTKR 1316
Query: 276 DLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLEI--EIREITIKL 449
D+ R + L ++L+ L +++ +E + KAD R+ + E + K
Sbjct: 1317 LADEESRERATLLGKFRNLEHDLDNLREQVEEEAEG--KADLQRQLSKANAEAQVWRSKY 1374
Query: 450 EEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFKELQQQAD-- 623
E + E + A +KLQ RL E E E+ ++C ++ + ++LQ + D
Sbjct: 1375 ESDGVARSEELEEAKRKLQARLAEAEETIESLNQKCIGLEKTKQRLSTEVEDLQLEVDRA 1434
Query: 624 -------------------DDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKY 746
+ + V +L LD +Q + R Y +L ++ Y
Sbjct: 1435 NAIANAAEKKQKAFDKIIGEWKLKVDDLAAELDASQKECRNYSTELFRLK-------GAY 1487
Query: 747 RKAQQQIE 770
+ Q+Q+E
Sbjct: 1488 EEGQEQLE 1495
>sp|P24733|MYS_AEQIR Myosin heavy chain, striated muscle Length = 1938 Score = 214 bits (545), Expect = 4e-55 Identities = 116/278 (41%), Positives = 179/278 (64%) Frame = +3 Query: 63 ESLLISERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKR 242 ES ++ER+ + GE+++LR LE +ERARK ++NE + RV+ELT V+++ KR Sbjct: 1666 ESYNMAERRCTLMSGEVEELRAALEQAERARKASDNELADANDRVNELTSQVSSVQGQKR 1725 Query: 243 RLEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLEI 422 +LEGD+ +Q DLD+ +++R K + RLADELR EQD+ + + +RK LE Sbjct: 1726 KLEGDINAMQTDLDEMHGELKGADERCKKAMADAARLADELRAEQDHSNQVEKVRKNLES 1785 Query: 423 EIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFK 602 +++E I+L+EAEA + + GK+ IQKL+ R+ ELE + + EQRR E RK +R+ K Sbjct: 1786 QVKEFQIRLDEAEASSLKGGKKMIQKLESRVHELEAELDNEQRRHAETQKNMRKADRRLK 1845 Query: 603 ELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEH 782 EL QAD+DR+ LQ+L+DK K++ +KRQ+EE E+++ + + KYRKAQ ++E+AE Sbjct: 1846 ELAFQADEDRKNQERLQELIDKLNAKIKTFKRQVEEAEEIAAINLAKYRKAQHELEEAEE 1905 Query: 783 RADMAERHLTVRRAGSVMPGSTYRAISVVRETSTVNTS 896 RAD A+ L RA S ++SV R + +V+ S Sbjct: 1906 RADTADSTLQKFRAKS------RSSVSVQRSSVSVSAS 1937
Score = 74.3 bits (181), Expect = 6e-13
Identities = 60/269 (22%), Positives = 121/269 (44%), Gaps = 28/269 (10%)
Frame = +3
Query: 81 ERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKRRLE--- 251
E L+ ++ DL N L+ +E+ + + +N+ L G +S+ H+ L +K+ LE
Sbjct: 941 EADNANLKKDIGDLENTLQKAEQDKAHKDNQISTLQGEISQQDEHIGKLNKEKKALEEAN 1000
Query: 252 --------------GDLGVIQGDLDDAVNARLASEDRANKLNNELNR----LADELRQEQ 377
L ++ L+ A++ + +R K+ ++ + + +L+ Q
Sbjct: 1001 KKTSDSLQAEEDKCNHLNKLKAKLEQALDELEDNLEREKKVRGDVEKAKRKVEQDLKSTQ 1060
Query: 378 DNYKKADSLRKTLE-------IEIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDF 536
+N + + +++ LE EI + KLE+ E + +R I++LQ R+ ELE +
Sbjct: 1061 ENVEDLERVKRELEENVRRKEAEISSLNSKLED-EQNLVSQLQRKIKELQARIEELEEEL 1119
Query: 537 EAEQRRCKEALAQARKFERQFKELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIE 716
EAE+ + Q + R+ +EL ++ D+ +L K + ++ +R LEE
Sbjct: 1120 EAERNARAKVEKQRAELNRELEELGERLDEAGGATSAQIELNKKREAELLKIRRDLEEAS 1179
Query: 717 DVSTVAMNKYRKAQQQIEDAEHRADMAER 803
++ RK Q + A AD ++
Sbjct: 1180 LQHEAQISALRKKHQ--DAANEMADQVDQ 1206
Score = 70.5 bits (171), Expect = 9e-12
Identities = 48/233 (20%), Positives = 112/233 (48%), Gaps = 1/233 (0%)
Frame = +3
Query: 96 QLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKRRLEGDLGVIQG 275
+++ +++ ++ + A R KN +E +L ++SE + L +RRLE + +Q
Sbjct: 1480 RIKASIEEYQDSIGALRRENKNLADEIHDLTDQLSEGGRSTHELDKARRRLEMEKEELQA 1539
Query: 276 DLDDAVNARLASEDRANKLNNELNRLADEL-RQEQDNYKKADSLRKTLEIEIREITIKLE 452
L++A A E + + E+ + +E+ ++ Q+ ++ D+ R+ + + + LE
Sbjct: 1540 ALEEAEGALEQEEAKVMRAQLEIATVRNEIDKRIQEKEEEFDNTRRNHQRALESMQASLE 1599
Query: 453 EAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFKELQQQADDDR 632
AEA + R +KL+ + ELE +A R E +++++Q +E+Q ++++
Sbjct: 1600 -AEAKGKADAMRIKKKLEQDINELEVALDASNRGKAEMEKTVKRYQQQIREMQTSIEEEQ 1658
Query: 633 RMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEHRAD 791
R E ++ + + + ++EE+ A + + ++ DA R +
Sbjct: 1659 RQRDEARESYNMAERRCTLMSGEVEELRAALEQAERARKASDNELADANDRVN 1711
Score = 68.9 bits (167), Expect = 2e-11
Identities = 63/257 (24%), Positives = 113/257 (43%), Gaps = 9/257 (3%)
Frame = +3
Query: 60 NESLLISE--RKRVQLQGELDDLRNQLEASERARKNAEN-------EAQELGGRVSELTL 212
+E L+S+ RK +LQ +++L +LEA AR E E +ELG R+ E
Sbjct: 1093 DEQNLVSQLQRKIKELQARIEELEEELEAERNARAKVEKQRAELNRELEELGERLDEAGG 1152
Query: 213 HVNTLINDKRRLEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKK 392
+ I ++ E +L I+ DL++A L E + + L + A+E+ + D +K
Sbjct: 1153 ATSAQIELNKKREAELLKIRRDLEEA---SLQHEAQISALRKKHQDAANEMADQVDQLQK 1209
Query: 393 ADSLRKTLEIEIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALA 572
S LE + +++ ++++ E+ T K + C E +
Sbjct: 1210 VKS---KLEKDKKDLKREMDDLESQMTHNMKN--------------------KGCSEKVM 1246
Query: 573 QARKFERQFKELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRK 752
++FE Q +L + +D +R + ELQ + Q + RQLE+ E +V + +
Sbjct: 1247 --KQFESQMSDLNARLEDSQRSINELQSQKSRLQAENSDLTRQLEDAEHRVSVLSKEKSQ 1304
Query: 753 AQQQIEDAEHRADMAER 803
Q+EDA + R
Sbjct: 1305 LSSQLEDARRSLEEETR 1321
Score = 63.5 bits (153), Expect = 1e-09
Identities = 62/286 (21%), Positives = 120/286 (41%), Gaps = 45/286 (15%)
Frame = +3
Query: 81 ERKRVQLQGELDDLR----------NQLEASERARKNAENEAQELGGRVSELTLHVNTLI 230
E+ + +LQ EL+D+ NQ+E +RA E Q +V+ L +
Sbjct: 1412 EKAKSRLQQELEDMSIEVDRANASVNQMEKKQRAFDKTTAEWQ---AKVNSLQSELENSQ 1468
Query: 231 NDKRRLEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRK 410
+ R +L I+ +++ ++ A L +E++ L D+L + + + D R+
Sbjct: 1469 KESRGYSAELYRIKASIEEYQDSIGALRRENKNLADEIHDLTDQLSEGGRSTHELDKARR 1528
Query: 411 TLEIEIREITIKLEEAEAYATREGKRAIQ----------KLQVRLRELETDF-------- 536
LE+E E+ LEEAE +E + ++ ++ R++E E +F
Sbjct: 1529 RLEMEKEELQAALEEAEGALEQEEAKVMRAQLEIATVRNEIDKRIQEKEEEFDNTRRNHQ 1588
Query: 537 ----------EAEQRRCKEALAQARKFERQFKELQQQADDDRRMVVELQDLLDKTQLKMR 686
EAE + +A+ +K E+ EL+ D R E++ + + Q ++R
Sbjct: 1589 RALESMQASLEAEAKGKADAMRIKKKLEQDINELEVALDASNRGKAEMEKTVKRYQQQIR 1648
Query: 687 AYKRQLEEIEDVSTVAMNKYRKAQQ-------QIEDAEHRADMAER 803
+ +EE + A Y A++ ++E+ + AER
Sbjct: 1649 EMQTSIEEEQRQRDEARESYNMAERRCTLMSGEVEELRAALEQAER 1694
Score = 62.8 bits (151), Expect = 2e-09
Identities = 52/246 (21%), Positives = 107/246 (43%), Gaps = 14/246 (5%)
Frame = +3
Query: 78 SERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTL-------HVNTLIND 236
SE+ Q + ++ DL +LE S+R+ +++ L S+LT V+ L +
Sbjct: 1242 SEKVMKQFESQMSDLNARLEDSQRSINELQSQKSRLQAENSDLTRQLEDAEHRVSVLSKE 1301
Query: 237 KRRLEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTL 416
K +L L + L++ AR ++ ++ +++ + ++L +EQ++
Sbjct: 1302 KSQLSSQLEDARRSLEEETRARSKLQNEVRNMHADMDAIREQLEEEQESKSDVQRQLSKA 1361
Query: 417 EIEIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQ 596
EI++ K E A T E + +KL +L E E EA +C + +++
Sbjct: 1362 NNEIQQWRSKFESEGANRTEELEDQKRKLLGKLSEAEQTTEAANAKCSALEKAKSRLQQE 1421
Query: 597 FKELQQQADDDRRMVVEL---QDLLDKT----QLKMRAYKRQLEEIEDVSTVAMNKYRKA 755
+++ + D V ++ Q DKT Q K+ + + +LE + S + +
Sbjct: 1422 LEDMSIEVDRANASVNQMEKKQRAFDKTTAEWQAKVNSLQSELENSQKESRGYSAELYRI 1481
Query: 756 QQQIED 773
+ IE+
Sbjct: 1482 KASIEE 1487
Score = 59.3 bits (142), Expect = 2e-08
Identities = 60/268 (22%), Positives = 106/268 (39%), Gaps = 19/268 (7%)
Frame = +3
Query: 75 ISERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKRRLEG 254
+ +++ QL +L+D R LE RAR +NE R +
Sbjct: 1297 VLSKEKSQLSSQLEDARRSLEEETRARSKLQNEV---------------------RNMHA 1335
Query: 255 DLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLEIEIRE 434
D+ I+ L++ ++ + + +K NNE+ + + E N + LE + R+
Sbjct: 1336 DMDAIREQLEEEQESKSDVQRQLSKANNEIQQWRSKFESEGAN------RTEELEDQKRK 1389
Query: 435 ITIKLEEAE--AYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFKEL 608
+ KL EAE A A++K + RL++ D E R ++ Q K +R F
Sbjct: 1390 LLGKLSEAEQTTEAANAKCSALEKAKSRLQQELEDMSIEVDRANASVNQMEKKQRAF--- 1446
Query: 609 QQQADDDRRMVVELQDLLDKTQLKMRAY-------KRQLEEIEDVSTVAMNKYRKAQQQI 767
+ + + V LQ L+ +Q + R Y K +EE +D + + +I
Sbjct: 1447 DKTTAEWQAKVNSLQSELENSQKESRGYSAELYRIKASIEEYQDSIGALRRENKNLADEI 1506
Query: 768 ED----------AEHRADMAERHLTVRR 821
D + H D A R L + +
Sbjct: 1507 HDLTDQLSEGGRSTHELDKARRRLEMEK 1534
>sp|Q90339|MYSS_CYPCA Myosin heavy chain, fast skeletal muscle Length = 1935 Score = 211 bits (537), Expect = 3e-54 Identities = 107/256 (41%), Positives = 170/256 (66%) Frame = +3 Query: 63 ESLLISERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKR 242 E + + ER+ +Q E+++LR LE +ER RK AE E + RV L +LIN K+ Sbjct: 1670 EQVAMVERRNSLMQAEIEELRAALEQTERGRKVAEQELVDASERVGLLHSQNTSLINTKK 1729 Query: 243 RLEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLEI 422 +LE DL +QG++DDAV +E++A K + +A+EL++EQD + ++K LE+ Sbjct: 1730 KLEADLVQVQGEVDDAVQEARNAEEKAKKAITDAAMMAEELKKEQDTSAHLERMKKNLEV 1789 Query: 423 EIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFK 602 ++++ +L+EAE+ A + GK+ +QKL+ R+RELE + EAEQRR +A+ RK+ER+ K Sbjct: 1790 TVKDLQHRLDEAESLAMKGGKKQLQKLESRVRELEAEVEAEQRRGADAVKGVRKYERRVK 1849 Query: 603 ELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEH 782 EL Q ++D++ V+ LQDL+DK QLK++ YKRQ EE E+ + +++YRK Q ++E+A+ Sbjct: 1850 ELTYQTEEDKKNVIRLQDLVDKLQLKVKVYKRQAEEAEEQTNTHLSRYRKVQHELEEAQE 1909 Query: 783 RADMAERHLTVRRAGS 830 RAD+AE + RA S Sbjct: 1910 RADVAESQVNKLRAKS 1925
Score = 65.5 bits (158), Expect = 3e-10
Identities = 62/283 (21%), Positives = 126/283 (44%), Gaps = 17/283 (6%)
Frame = +3
Query: 81 ERKRVQLQGELDDLRNQLEAS-------ERARKNAENEAQELGGRVSELTLHVNTLINDK 239
E+ + +LQGE++DL +E + ++ ++N + E + E + +
Sbjct: 1416 EKTKQRLQGEVEDLMIDVERANSLAANLDKKQRNFDKVLAEWKQKYEESQAELEGAQKEA 1475
Query: 240 RRLEGDLGVIQGDLDDAVNARLASEDRANK-LNNELNRLADELRQEQDNYKKADSLRKTL 416
R L +L ++ ++A++ L + R NK L E++ L ++L + + + + +KT+
Sbjct: 1476 RSLSTELFKMKNSYEEALD-HLETLKRENKNLQQEISDLTEQLGETGKSIHELEKAKKTV 1534
Query: 417 EIEIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQ-ARKFER 593
E E EI LEEAE E + I ++Q+ L +++++ + + E + Q R +R
Sbjct: 1535 ESEKSEIQTALEEAEGTLEHEESK-ILRVQLELNQVKSEIDRKLAEKDEEMEQIKRNSQR 1593
Query: 594 QFKELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIED 773
+Q D + R + + K + + + QL + A + R Q Q++D
Sbjct: 1594 VIDSMQSTLDSEVRSRNDALRVKKKMEGDLNEMEIQLSHANRQAAEAQKQLRNVQGQLKD 1653
Query: 774 AE--------HRADMAERHLTVRRAGSVMPGSTYRAISVVRET 878
A+ + DM E+ V R S+M + + +T
Sbjct: 1654 AQLHLDEAVRGQEDMKEQVAMVERRNSLMQAEIEELRAALEQT 1696
Score = 63.2 bits (152), Expect = 1e-09
Identities = 69/256 (26%), Positives = 120/256 (46%), Gaps = 23/256 (8%)
Frame = +3
Query: 72 LISERKRVQL--QGELDDLR---NQLEASERARKNAENEAQELGGRVSELTLHVNTLIND 236
L E+K +Q Q LDDL+ +++ +A+ E + +L G + + L
Sbjct: 993 LTKEKKALQEAHQQTLDDLQAEEDKVNTLTKAKTKLEQQVDDLEGSLEQEKKLRMDLERA 1052
Query: 237 KRRLEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTL 416
KR+LEGDL + Q + D N + S+++ K + E+++L ++ EQ + K L
Sbjct: 1053 KRKLEGDLKLAQESIMDLENEKQQSDEKIKKKDFEISQLLSKIEDEQSLGAQLQKKIKEL 1112
Query: 417 EIEIREITIKLE-EAEAYATREGKRA-----IQKLQVRLRELETDFEAEQRRCKEALAQA 578
+ I E+ ++E E A A E +RA ++++ RL E A+ K+ A+
Sbjct: 1113 QARIEELEEEIEAERAARAKVEKQRADLSRELEEISERLEEAGGATAAQIEMNKKREAEF 1172
Query: 579 RKFERQFKE--LQQQA------DDDRRMVVELQDLLDKTQ-LKMRAYKRQLE---EIEDV 722
+K R +E LQ +A + V EL + +D Q +K + K + E EI+D+
Sbjct: 1173 QKMRRDLEESTLQHEATAAALRKEQADSVAELGEQIDNLQRVKQKLEKEKSEYKMEIDDL 1232
Query: 723 STVAMNKYRKAQQQIE 770
T M KA+ +E
Sbjct: 1233 -TSNMEAVAKAKANLE 1247
Score = 57.8 bits (138), Expect = 6e-08
Identities = 61/288 (21%), Positives = 115/288 (39%)
Frame = +3
Query: 84 RKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKRRLEGDLG 263
+ ++QL+ +L + +LE E NAE A+ KR+LE +
Sbjct: 911 KSKIQLEAKLKETNERLEDEEEI--NAELTAK-------------------KRKLEDECS 949
Query: 264 VIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLEIEIREITI 443
++ D+DD E + N++ L +E+ + ++ K +K L+ E + T+
Sbjct: 950 ELKKDIDDLELTLAKVEKEKHATENKVKNLTEEMASQDESIAKLTKEKKALQ-EAHQQTL 1008
Query: 444 KLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFKELQQQAD 623
+AE +A KL+ ++ +LE E E++ + RK E K Q+
Sbjct: 1009 DDLQAEEDKVNTLTKAKTKLEQQVDDLEGSLEQEKKLRMDLERAKRKLEGDLKLAQESIM 1068
Query: 624 DDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEHRADMAER 803
D + + + K ++ ++E+ + + K ++ Q +IE+ E + AER
Sbjct: 1069 DLENEKQQSDEKIKKKDFEISQLLSKIEDEQSLGAQLQKKIKELQARIEELEEEIE-AER 1127
Query: 804 HLTVRRAGSVMPGSTYRAISVVRETSTVNTSRGARATSIR*TKSNEKE 947
R R + + E + + GA A I K E E
Sbjct: 1128 --AARAKVEKQRADLSRELEEISE--RLEEAGGATAAQIEMNKKREAE 1171
Score = 50.1 bits (118), Expect = 1e-05
Identities = 48/244 (19%), Positives = 105/244 (43%), Gaps = 7/244 (2%)
Frame = +3
Query: 81 ERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTL-------INDK 239
++K +LQ +++L ++EA AR E + +L + E++ + I
Sbjct: 1106 QKKIKELQARIEELEEEIEAERAARAKVEKQRADLSRELEEISERLEEAGGATAAQIEMN 1165
Query: 240 RRLEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTLE 419
++ E + ++ DL+++ L E A L E EL ++ DN ++ +++ LE
Sbjct: 1166 KKREAEFQKMRRDLEEST---LQHEATAAALRKEQADSVAELGEQIDNLQR---VKQKLE 1219
Query: 420 IEIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQF 599
E E +++++ + A+ K + L ++ R E Q
Sbjct: 1220 KEKSEYKMEIDDLTS-----NMEAVAKAKANLEKM-----------------CRTLEDQL 1257
Query: 600 KELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAE 779
E++ ++D++ R + ++ + Q + + RQLEE E + + + QQIE+ +
Sbjct: 1258 SEIKTKSDENVRQLNDMNAQRARLQTENGEFSRQLEEKEALVSQLTRGKQAYTQQIEELK 1317
Query: 780 HRAD 791
+
Sbjct: 1318 RHIE 1321
>sp|P02566|MYO4_CAEEL Myosin-4 (Myosin heavy chain B) (MHC B) (Uncoordinated protein 54) Length = 1966 Score = 209 bits (533), Expect = 9e-54 Identities = 115/286 (40%), Positives = 181/286 (63%), Gaps = 6/286 (2%) Frame = +3 Query: 57 TNESLLISERKRVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLIND 236 T E +E++ LQ E ++L EA+ERARK AE EA + + +E V++L + Sbjct: 1679 TREQFFNAEKRATLLQSEKEELLVANEAAERARKQAEYEAADARDQANEANAQVSSLTSA 1738 Query: 237 KRRLEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRLADELRQEQDNYKKADSLRKTL 416 KR+LEG++ I DLD+ +N A+E+R+ K + RLA+ELRQEQ++ + D LRK L Sbjct: 1739 KRKLEGEIQAIHADLDETLNEYKAAEERSKKAIADATRLAEELRQEQEHSQHVDRLRKGL 1798 Query: 417 EIEIREITIKLEEAEAYATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQ 596 E +++EI ++L+EAEA A + GK+ I KL+ R+RELE++ + EQRR ++A + +R+ Sbjct: 1799 EQQLKEIQVRLDEAEAAALKGGKKVIAKLEQRVRELESELDGEQRRFQDANKNLGRADRR 1858 Query: 597 FKELQQQADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDA 776 +ELQ Q D+D++ LQDL+DK Q K++ K+Q+EE E+++ + + KY++ Q+EDA Sbjct: 1859 VRELQFQVDEDKKNFERLQDLIDKLQQKLKTQKKQVEEAEELANLNLQKYKQLTHQLEDA 1918 Query: 777 EHRADMAERHLT-----VRRAGSVMPG-STYRAISVVRETSTVNTS 896 E RAD AE L+ R + SV PG + + +V+R S S Sbjct: 1919 EERADQAENSLSKMRSKSRASASVAPGLQSSASAAVIRSPSRARAS 1964
Score = 63.9 bits (154), Expect = 8e-10
Identities = 50/234 (21%), Positives = 110/234 (47%), Gaps = 1/234 (0%)
Frame = +3
Query: 114 DDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKRRLEGDLGVIQGDLDDAV 293
++L +E R K+ E ++L ++ E V+ + RRLE + +Q LD+A
Sbjct: 1501 EELAEVVEGLRRENKSLSQEIKDLTDQLGEGGRSVHEMQKIIRRLEIEKEELQHALDEAE 1560
Query: 294 NARLASEDRANKLNNELNRLADELRQE-QDNYKKADSLRKTLEIEIREITIKLEEAEAYA 470
A A E + + E++++ E+ + Q+ ++ ++ RK + + LE EA
Sbjct: 1561 AALEAEESKVLRAQVEVSQIRSEIEKRIQEKEEEFENTRKNHARALESMQASLE-TEAKG 1619
Query: 471 TREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFKELQQQADDDRRMVVEL 650
E R +KL+ + ELE + + +A ++++ Q +ELQ Q ++++R +
Sbjct: 1620 KAELLRIKKKLEGDINELEIALDHANKANADAQKNLKRYQEQVRELQLQVEEEQRNGADT 1679
Query: 651 QDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEHRADMAERHLT 812
++ + + + + EE+ + A ++A+ + DA +A+ A ++
Sbjct: 1680 REQFFNAEKRATLLQSEKEELLVANEAAERARKQAEYEAADARDQANEANAQVS 1733
Score = 60.8 bits (146), Expect = 7e-09
Identities = 64/302 (21%), Positives = 124/302 (41%), Gaps = 64/302 (21%)
Frame = +3
Query: 90 RVQLQGELDDLRNQLEASERARKNAENEAQELGGRVSELTLHVNTLINDKRRLEGDLGVI 269
+ +L+ LDDL + LE +RAR + + + +++ G + +++ + LE +L
Sbjct: 1036 KAKLEQTLDDLEDSLEREKRARADLDKQKRKVEGELKIAQENIDESGRQRHDLENNLKKK 1095
Query: 270 QGDLDDAVNARLASEDR-ANKLNNEL-------NRLADELRQEQDNYKKADSLRKTLEIE 425
+ +L +V++RL E +KL ++ + L +EL E+ + KAD + L+ E
Sbjct: 1096 ESELH-SVSSRLEDEQALVSKLQRQIKDGQSRISELEEELENERQSRSKADRAKSDLQRE 1154
Query: 426 IREITIKLEE------AEAYATREGKRAIQKLQVRLRELETDFEAE----QRRCKEALAQ 575
+ E+ KL+E A+ ++ + + KL+ L E + E + +++ +A+A+
Sbjct: 1155 LEELGEKLDEQGGATAAQVEVNKKREAELAKLRRDLEEANMNHENQLGGLRKKHTDAVAE 1214
Query: 576 ----------------------------------------------ARKFERQFKELQQQ 617
A++FE Q ELQ +
Sbjct: 1215 LTDQLDQLNKAKAKVEKDKAQAVRDAEDLAAQLDQETSGKLNNEKLAKQFELQLTELQSK 1274
Query: 618 ADDDRRMVVELQDLLDKTQLKMRAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEHRADMA 797
AD+ R + + L + + RQLE+ E + Q+E+A AD
Sbjct: 1275 ADEQSRQLQDFTSLKGRLHSENGDLVRQLEDAESQVNQLTRLKSQLTSQLEEARRTADEE 1334
Query: 798 ER 803
R
Sbjct: 1335 AR 1336
Score = 48.5 bits (114), Expect = 3e-05
Identities = 61/307 (19%), Positives = 120/307 (39%), Gaps = 64/307 (20%)
Frame = +3
Query: 81 ERKRVQLQGELDDLRNQLEASE----RARKNAENEAQELGGRVSE--------------- 203
E ++ +LQ LD+ LEA E RA+ E+ R+ E
Sbjct: 1546 EIEKEELQHALDEAEAALEAEESKVLRAQVEVSQIRSEIEKRIQEKEEEFENTRKNHARA 1605
Query: 204 ---LTLHVNT-------LINDKRRLEGDLGVIQGDLDDAVNARLASEDRANKLNNELNRL 353
+ + T L+ K++LEGD+ ++ LD A A ++ + ++ L
Sbjct: 1606 LESMQASLETEAKGKAELLRIKKKLEGDINELEIALDHANKANADAQKNLKRYQEQVREL 1665
Query: 354 ADELRQEQDN----------------------------YKKADSLRKTLEIEIREITIKL 449
++ +EQ N + A+ RK E E + +
Sbjct: 1666 QLQVEEEQRNGADTREQFFNAEKRATLLQSEKEELLVANEAAERARKQAEYEAADARDQA 1725
Query: 450 EEAEAY------ATREGKRAIQKLQVRLRELETDFEAEQRRCKEALAQARKFERQFKELQ 611
EA A A R+ + IQ + L E +++A + R K+A+A A + + ++ Q
Sbjct: 1726 NEANAQVSSLTSAKRKLEGEIQAIHADLDETLNEYKAAEERSKKAIADATRLAEELRQEQ 1785
Query: 612 QQADDDRRMVVELQDLLDKTQLKM-RAYKRQLEEIEDVSTVAMNKYRKAQQQIEDAEHRA 788
+ + R+ L+ L + Q+++ A L+ + V + R+ + +++ + R
Sbjct: 1786 EHSQHVDRLRKGLEQQLKEIQVRLDEAEAAALKGGKKVIAKLEQRVRELESELDGEQRRF 1845
Query: 789 DMAERHL 809
A ++L
Sbjct: 1846 QDANKNL 1852
Score = 38.1 bits (87), Expect = 0.047
Identities = 67/294 (22%), Positives = 111/294 (37%), Gaps = 63/294 (21%)
Frame = +3
Query: 81 ERKRVQLQGELDDLRNQLEA-------SERARKNAENEAQELGGRVSELTLHVNTLINDK 239
E+ + Q + +DL QL+ +E+ K E + EL + E + + + K
Sbjct: 1230 EKDKAQAVRDAEDLAAQLDQETSGKLNNEKLAKQFELQLTELQSKADEQSRQLQDFTSLK 1289
Query: 240 RRLEGDLGVIQGDLDDA---VNARLASEDRANKLNNELNRLADELRQEQD-------NYK 389
RL + G + L+DA VN + + E R ADE +E+ NY+
Sbjct: 1290 GRLHSENGDLVRQLEDAESQVNQLTRLKSQLTSQLEEARRTADEEARERQTVAAQAKNYQ 1349
Query: 390 -KADSLRKTLEIEIR---EITIKLEEAEAY---------------------ATREGKRAI 494
+A+ L+++LE EI EI +L +A A A R + I
Sbjct: 1350 HEAEQLQESLEEEIEGKNEILRQLSKANADIQQWKARFEGEGLLKADELEDAKRRQAQKI 1409
Query: 495 QKLQVRL--------------RELETDFEAEQRRCKEALAQARKFERQFKELQQQADDDR 632
+LQ L L D + Q + A A E++ K + D+ R
Sbjct: 1410 NELQEALDAANSKNASLEKTKSRLVGDLDDAQVDVERANGVASALEKKQKGFDKIIDEWR 1469
Query: 633 RMVVELQDLLDKTQLKMRAYKRQL-------EEIEDVSTVAMNKYRKAQQQIED 773
+ +L LD Q +R L EE+ +V + + Q+I+D
Sbjct: 1470 KKTDDLAAELDGAQRDLRNTSTDLFKAKNAQEELAEVVEGLRRENKSLSQEIKD 1523
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 93,017,956
Number of Sequences: 369166
Number of extensions: 1546571
Number of successful extensions: 9170
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6687
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8292
length of database: 68,354,980
effective HSP length: 112
effective length of database: 47,664,660
effective search space used: 12678799560
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail