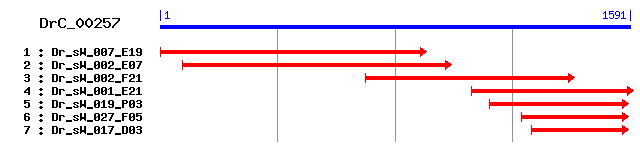
DrC_00257
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00257 (1591 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P81178|ALDH2_MESAU Aldehyde dehydrogenase, mitochondrial... 557 e-158 sp|P05091|ALDH2_HUMAN Aldehyde dehydrogenase, mitochondrial... 554 e-157 sp|P11884|ALDH2_RAT Aldehyde dehydrogenase, mitochondrial p... 554 e-157 sp|P47738|ALDH2_MOUSE Aldehyde dehydrogenase, mitochondrial... 550 e-156 sp|P12762|ALDH2_HORSE Aldehyde dehydrogenase, mitochondrial... 548 e-155 sp|Q63639|AL1A2_RAT Retinal dehydrogenase 2 (RalDH2) (RALDH... 547 e-155 sp|Q62148|AL1A2_MOUSE Retinal dehydrogenase 2 (RalDH2) (RAL... 547 e-155 sp|P20000|ALDH2_BOVIN Aldehyde dehydrogenase, mitochondrial... 547 e-155 sp|O94788|AL1A2_HUMAN Retinal dehydrogenase 2 (RalDH2) (RAL... 543 e-154 sp|P27463|AL1A1_CHICK Retinal dehydrogenase 1 (RalDH1) (RAL... 540 e-153
>sp|P81178|ALDH2_MESAU Aldehyde dehydrogenase, mitochondrial (ALDH class 2) (ALDH1) (ALDH-E2) Length = 500 Score = 557 bits (1435), Expect = e-158 Identities = 279/486 (57%), Positives = 345/486 (70%), Gaps = 1/486 (0%) Frame = +2 Query: 65 PEIKFTKIFINNEWCNSEEGLTFPTINPTNEQVIVQVQEGREXXXXXXXXXXXXXXXXGS 244 PE+ +IFINNEW ++ TFPT+NP+ +VI QV EG + GS Sbjct: 15 PEVFCNQIFINNEWHDAVSKKTFPTVNPSTGEVICQVAEGSKEDVDKAVKAARAAFQLGS 74 Query: 245 EWRTLNASARGLLLNKLADLIELEADYISRLEALDNGKSYVIAK-GDIEFSVKILRYYAG 421 WR ++AS RG LLN+LADLIE + Y++ LE LDNGK YVI+ D++ +K LRYYAG Sbjct: 75 PWRRMDASDRGRLLNRLADLIERDRTYLAALETLDNGKPYVISYLVDLDMVLKCLRYYAG 134 Query: 422 YADKIHGVTTAPDGDFLSYTRIEPVGVVGGIIPWNYPFMLAIFKVGPALAAGCTLVLKPA 601 +ADK HG T DGDF SYTR EPVGV G IIPWN+P ++ +K+GPALA G +V+K A Sbjct: 135 WADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQAWKLGPALATGNVVVMKVA 194 Query: 602 EQTPLSALFLGDLILRAGFPKGVVNIVPGYGPTAGAALSSHHDVRKITFTGSTEIGHVIM 781 EQTPL+AL++ +LI AGFP GVVNIVPG+GPTAGAA++SH DV K+ FTGSTE+GH+I Sbjct: 195 EQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASHEDVDKVAFTGSTEVGHLIQ 254 Query: 782 KTIGETSLKRVTLELGGKSPLIIFXXXXXXXXXXXXXXXCMTNHGQCCVAGTRTFVEASI 961 G ++LKRVTLELGGKSP II N GQCC AG+RTFV+ + Sbjct: 255 VAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALFFNQGQCCCAGSRTFVQEDV 314 Query: 962 YDTIIEKLKKLAEERIVGDPFDEKTIQGPQVDKEQFNKILDLIESGKHEGAKLVTGGNRI 1141 YD +E+ A+ R+VG+PFD +T QGPQVD+ QF KIL I+SG+ EGAKL+ GG Sbjct: 315 YDEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKILGYIKSGQQEGAKLLCGGGAA 374 Query: 1142 GNVGYFIQPTVFGDVKDNMRIAKEEIFGPVQSVFKFNTMEEVIERANNSDYGLGAGVFTS 1321 + GYFIQPTVFGDVKD M IAKEEIFGPV + KF T+EEV+ RANNS YGL A VFT Sbjct: 375 ADRGYFIQPTVFGDVKDGMTIAKEEIFGPVMQILKFKTIEEVVGRANNSKYGLAAAVFTK 434 Query: 1322 DINKAIMMTQALEAGSVWVNCYDVGLVTAPFGGFKQSGVGRELGCYGLEPYIEVKGVTIA 1501 D++KA ++QAL+AG+VW+NCYDV +PFGG+K SG GRELG YGL+ Y EVK VTI Sbjct: 435 DLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGSGRELGEYGLQAYTEVKTVTIK 494 Query: 1502 IPQKNS 1519 +PQKNS Sbjct: 495 VPQKNS 500
>sp|P05091|ALDH2_HUMAN Aldehyde dehydrogenase, mitochondrial precursor (ALDH class 2) (ALDHI) (ALDH-E2) Length = 517 Score = 554 bits (1428), Expect = e-157 Identities = 281/486 (57%), Positives = 343/486 (70%), Gaps = 1/486 (0%) Frame = +2 Query: 65 PEIKFTKIFINNEWCNSEEGLTFPTINPTNEQVIVQVQEGREXXXXXXXXXXXXXXXXGS 244 PE+ +IFINNEW ++ TFPT+NP+ +VI QV EG + GS Sbjct: 32 PEVFCNQIFINNEWHDAVSRKTFPTVNPSTGEVICQVAEGDKEDVDKAVKAARAAFQLGS 91 Query: 245 EWRTLNASARGLLLNKLADLIELEADYISRLEALDNGKSYVIAK-GDIEFSVKILRYYAG 421 WR ++AS RG LLN+LADLIE + Y++ LE LDNGK YVI+ D++ +K LRYYAG Sbjct: 92 PWRRMDASHRGRLLNRLADLIERDRTYLAALETLDNGKPYVISYLVDLDMVLKCLRYYAG 151 Query: 422 YADKIHGVTTAPDGDFLSYTRIEPVGVVGGIIPWNYPFMLAIFKVGPALAAGCTLVLKPA 601 +ADK HG T DGDF SYTR EPVGV G IIPWN+P ++ +K+GPALA G +V+K A Sbjct: 152 WADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQAWKLGPALATGNVVVMKVA 211 Query: 602 EQTPLSALFLGDLILRAGFPKGVVNIVPGYGPTAGAALSSHHDVRKITFTGSTEIGHVIM 781 EQTPL+AL++ +LI AGFP GVVNIVPG+GPTAGAA++SH DV K+ FTGSTEIG VI Sbjct: 212 EQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASHEDVDKVAFTGSTEIGRVIQ 271 Query: 782 KTIGETSLKRVTLELGGKSPLIIFXXXXXXXXXXXXXXXCMTNHGQCCVAGTRTFVEASI 961 G ++LKRVTLELGGKSP II N GQCC AG+RTFV+ I Sbjct: 272 VAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALFFNQGQCCCAGSRTFVQEDI 331 Query: 962 YDTIIEKLKKLAEERIVGDPFDEKTIQGPQVDKEQFNKILDLIESGKHEGAKLVTGGNRI 1141 YD +E+ A+ R+VG+PFD KT QGPQVD+ QF KIL I +GK EGAKL+ GG Sbjct: 332 YDEFVERSVARAKSRVVGNPFDSKTEQGPQVDETQFKKILGYINTGKQEGAKLLCGGGIA 391 Query: 1142 GNVGYFIQPTVFGDVKDNMRIAKEEIFGPVQSVFKFNTMEEVIERANNSDYGLGAGVFTS 1321 + GYFIQPTVFGDV+D M IAKEEIFGPV + KF T+EEV+ RANNS YGL A VFT Sbjct: 392 ADRGYFIQPTVFGDVQDGMTIAKEEIFGPVMQILKFKTIEEVVGRANNSTYGLAAAVFTK 451 Query: 1322 DINKAIMMTQALEAGSVWVNCYDVGLVTAPFGGFKQSGVGRELGCYGLEPYIEVKGVTIA 1501 D++KA ++QAL+AG+VWVNCYDV +PFGG+K SG GRELG YGL+ Y EVK VT+ Sbjct: 452 DLDKANYLSQALQAGTVWVNCYDVFGAQSPFGGYKMSGSGRELGEYGLQAYTEVKTVTVK 511 Query: 1502 IPQKNS 1519 +PQKNS Sbjct: 512 VPQKNS 517
>sp|P11884|ALDH2_RAT Aldehyde dehydrogenase, mitochondrial precursor (ALDH class 2) (ALDH1) (ALDH-E2) Length = 519 Score = 554 bits (1427), Expect = e-157 Identities = 277/486 (56%), Positives = 344/486 (70%), Gaps = 1/486 (0%) Frame = +2 Query: 65 PEIKFTKIFINNEWCNSEEGLTFPTINPTNEQVIVQVQEGREXXXXXXXXXXXXXXXXGS 244 PE+ +IFINNEW ++ TFPT+NP+ +VI QV EG + GS Sbjct: 34 PEVFCNQIFINNEWHDAVSKKTFPTVNPSTGEVICQVAEGNKEDVDKAVKAAQAAFQLGS 93 Query: 245 EWRTLNASARGLLLNKLADLIELEADYISRLEALDNGKSYVIAK-GDIEFSVKILRYYAG 421 WR ++AS RG LL +LADLIE + Y++ LE LDNGK YVI+ D++ +K LRYYAG Sbjct: 94 PWRRMDASDRGRLLYRLADLIERDRTYLAALETLDNGKPYVISYLVDLDMVLKCLRYYAG 153 Query: 422 YADKIHGVTTAPDGDFLSYTRIEPVGVVGGIIPWNYPFMLAIFKVGPALAAGCTLVLKPA 601 +ADK HG T DGDF SYTR EPVGV G IIPWN+P ++ +K+GPALA G +V+K A Sbjct: 154 WADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQAWKLGPALATGNVVVMKVA 213 Query: 602 EQTPLSALFLGDLILRAGFPKGVVNIVPGYGPTAGAALSSHHDVRKITFTGSTEIGHVIM 781 EQTPL+AL++ +LI AGFP GVVNIVPG+GPTAGAA++SH DV K+ FTGSTE+GH+I Sbjct: 214 EQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASHEDVDKVAFTGSTEVGHLIQ 273 Query: 782 KTIGETSLKRVTLELGGKSPLIIFXXXXXXXXXXXXXXXCMTNHGQCCVAGTRTFVEASI 961 G ++LKRVTLELGGKSP II N GQCC AG+RTFV+ + Sbjct: 274 VAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALFFNQGQCCCAGSRTFVQEDV 333 Query: 962 YDTIIEKLKKLAEERIVGDPFDEKTIQGPQVDKEQFNKILDLIESGKHEGAKLVTGGNRI 1141 YD +E+ A+ R+VG+PFD +T QGPQVD+ QF KIL I+SG+ EGAKL+ GG Sbjct: 334 YDEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKILGYIKSGQQEGAKLLCGGGAA 393 Query: 1142 GNVGYFIQPTVFGDVKDNMRIAKEEIFGPVQSVFKFNTMEEVIERANNSDYGLGAGVFTS 1321 + GYFIQPTVFGDVKD M IAKEEIFGPV + KF T+EEV+ RANNS YGL A VFT Sbjct: 394 ADRGYFIQPTVFGDVKDGMTIAKEEIFGPVMQILKFKTIEEVVGRANNSKYGLAAAVFTK 453 Query: 1322 DINKAIMMTQALEAGSVWVNCYDVGLVTAPFGGFKQSGVGRELGCYGLEPYIEVKGVTIA 1501 D++KA ++QAL+AG+VW+NCYDV +PFGG+K SG GRELG YGL+ Y EVK VT+ Sbjct: 454 DLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGSGRELGEYGLQAYTEVKTVTVK 513 Query: 1502 IPQKNS 1519 +PQKNS Sbjct: 514 VPQKNS 519
>sp|P47738|ALDH2_MOUSE Aldehyde dehydrogenase, mitochondrial precursor (ALDH class 2) (AHD-M1) (ALDHI) (ALDH-E2) Length = 519 Score = 550 bits (1416), Expect = e-156 Identities = 275/486 (56%), Positives = 344/486 (70%), Gaps = 1/486 (0%) Frame = +2 Query: 65 PEIKFTKIFINNEWCNSEEGLTFPTINPTNEQVIVQVQEGREXXXXXXXXXXXXXXXXGS 244 PE+ +IFINNEW ++ TFPT+NP+ +VI QV EG + GS Sbjct: 34 PEVFCNQIFINNEWHDAVSRKTFPTVNPSTGEVICQVAEGNKEDVDKAVKAARAAFQLGS 93 Query: 245 EWRTLNASARGLLLNKLADLIELEADYISRLEALDNGKSYVIAK-GDIEFSVKILRYYAG 421 WR ++AS RG LL +LADLIE + Y++ LE LDNGK YVI+ D++ +K LRYYAG Sbjct: 94 PWRRMDASDRGRLLYRLADLIERDRTYLAALETLDNGKPYVISYLVDLDMVLKCLRYYAG 153 Query: 422 YADKIHGVTTAPDGDFLSYTRIEPVGVVGGIIPWNYPFMLAIFKVGPALAAGCTLVLKPA 601 +ADK HG T DGDF SYTR EPVGV G IIPWN+P ++ +K+GPALA G +V+K A Sbjct: 154 WADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQAWKLGPALATGNVVVMKVA 213 Query: 602 EQTPLSALFLGDLILRAGFPKGVVNIVPGYGPTAGAALSSHHDVRKITFTGSTEIGHVIM 781 EQTPL+AL++ +LI AGFP GVVNIVPG+GPTAGAA++SH V K+ FTGSTE+GH+I Sbjct: 214 EQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASHEGVDKVAFTGSTEVGHLIQ 273 Query: 782 KTIGETSLKRVTLELGGKSPLIIFXXXXXXXXXXXXXXXCMTNHGQCCVAGTRTFVEASI 961 G ++LKRVTLELGGKSP II N GQCC AG+RTFV+ ++ Sbjct: 274 VAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALFFNQGQCCCAGSRTFVQENV 333 Query: 962 YDTIIEKLKKLAEERIVGDPFDEKTIQGPQVDKEQFNKILDLIESGKHEGAKLVTGGNRI 1141 YD +E+ A+ R+VG+PFD +T QGPQVD+ QF KIL I+SG+ EGAKL+ GG Sbjct: 334 YDEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKILGYIKSGQQEGAKLLCGGGAA 393 Query: 1142 GNVGYFIQPTVFGDVKDNMRIAKEEIFGPVQSVFKFNTMEEVIERANNSDYGLGAGVFTS 1321 + GYFIQPTVFGDVKD M IAKEEIFGPV + KF T+EEV+ RAN+S YGL A VFT Sbjct: 394 ADRGYFIQPTVFGDVKDGMTIAKEEIFGPVMQILKFKTIEEVVGRANDSKYGLAAAVFTK 453 Query: 1322 DINKAIMMTQALEAGSVWVNCYDVGLVTAPFGGFKQSGVGRELGCYGLEPYIEVKGVTIA 1501 D++KA ++QAL+AG+VW+NCYDV +PFGG+K SG GRELG YGL+ Y EVK VT+ Sbjct: 454 DLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGSGRELGEYGLQAYTEVKTVTVK 513 Query: 1502 IPQKNS 1519 +PQKNS Sbjct: 514 VPQKNS 519
>sp|P12762|ALDH2_HORSE Aldehyde dehydrogenase, mitochondrial (ALDH class 2) (ALDHI) (ALDH-E2) Length = 500 Score = 548 bits (1413), Expect = e-155 Identities = 274/486 (56%), Positives = 343/486 (70%), Gaps = 1/486 (0%) Frame = +2 Query: 65 PEIKFTKIFINNEWCNSEEGLTFPTINPTNEQVIVQVQEGREXXXXXXXXXXXXXXXXGS 244 PE+ + +IFINNEW ++ TFPT+NP+ +VI QV G + GS Sbjct: 15 PEVFYNQIFINNEWHDAVSKKTFPTVNPSTGEVICQVAAGDKEDVDRAVKAARAAFQLGS 74 Query: 245 EWRTLNASARGLLLNKLADLIELEADYISRLEALDNGKSYVIAK-GDIEFSVKILRYYAG 421 WR ++AS RG LLN+LADLIE + Y++ LE LDNGK YVI+ D++ +K LRYYAG Sbjct: 75 PWRRMDASDRGRLLNRLADLIERDRTYLAALETLDNGKPYVISYLVDLDMVLKCLRYYAG 134 Query: 422 YADKIHGVTTAPDGDFLSYTRIEPVGVVGGIIPWNYPFMLAIFKVGPALAAGCTLVLKPA 601 +ADK HG T DGDF SYTR EPVGV G IIPWN+P ++ K+GPALA G +V+K A Sbjct: 135 WADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQAAKLGPALATGNVVVMKVA 194 Query: 602 EQTPLSALFLGDLILRAGFPKGVVNIVPGYGPTAGAALSSHHDVRKITFTGSTEIGHVIM 781 EQTPL+AL++ +L AGFP GVVN+VPG+GPTAGAA++SH DV K+ FTGSTE+GH+I Sbjct: 195 EQTPLTALYVANLTKEAGFPPGVVNVVPGFGPTAGAAIASHEDVDKVAFTGSTEVGHLIQ 254 Query: 782 KTIGETSLKRVTLELGGKSPLIIFXXXXXXXXXXXXXXXCMTNHGQCCVAGTRTFVEASI 961 G ++LK+VTLELGGKSP II N GQCC AG+RTFV+ + Sbjct: 255 VAAGRSNLKKVTLELGGKSPNIIVSDADMDWAVEQAHFALFFNQGQCCGAGSRTFVQEDV 314 Query: 962 YDTIIEKLKKLAEERIVGDPFDEKTIQGPQVDKEQFNKILDLIESGKHEGAKLVTGGNRI 1141 Y +E+ A+ R+VG+PFD +T QGPQVD+ QFNK+L I+SGK EGAKL+ GG Sbjct: 315 YAEFVERSVARAKSRVVGNPFDSQTEQGPQVDETQFNKVLGYIKSGKEEGAKLLCGGGAA 374 Query: 1142 GNVGYFIQPTVFGDVKDNMRIAKEEIFGPVQSVFKFNTMEEVIERANNSDYGLGAGVFTS 1321 + GYFIQPTVFGDV+D M IAKEEIFGPV + KF T+EEV+ RANNS YGL A VFT Sbjct: 375 ADRGYFIQPTVFGDVQDGMTIAKEEIFGPVMQILKFKTIEEVVGRANNSKYGLAAAVFTK 434 Query: 1322 DINKAIMMTQALEAGSVWVNCYDVGLVTAPFGGFKQSGVGRELGCYGLEPYIEVKGVTIA 1501 D++KA ++QAL+AG+VW+NCYDV +PFGG+K SG GRELG YGL+ Y EVK VTI Sbjct: 435 DLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGNGRELGEYGLQAYTEVKTVTIK 494 Query: 1502 IPQKNS 1519 +PQKNS Sbjct: 495 VPQKNS 500
>sp|Q63639|AL1A2_RAT Retinal dehydrogenase 2 (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2) Length = 499 Score = 547 bits (1410), Expect = e-155 Identities = 271/485 (55%), Positives = 349/485 (71%), Gaps = 1/485 (0%) Frame = +2 Query: 68 EIKFTKIFINNEWCNSEEGLTFPTINPTNEQVIVQVQEGREXXXXXXXXXXXXXXXXGSE 247 EIK+TKIFINNEW NSE G FP NP + + +VQE + GS Sbjct: 15 EIKYTKIFINNEWQNSESGRVFPVCNPATGEQVCEVQEADKVDIDKAVQAARLAFSLGSV 74 Query: 248 WRTLNASARGLLLNKLADLIELEADYISRLEALDNGKSYVIAKG-DIEFSVKILRYYAGY 424 WR ++AS RG LL+KLADL+E + ++ +E+L+ GK ++ A D++ +K LRYYAG+ Sbjct: 75 WRRMDASERGRLLDKLADLVERDRATLATMESLNGGKPFLQAFYIDLQGVIKTLRYYAGW 134 Query: 425 ADKIHGVTTAPDGDFLSYTRIEPVGVVGGIIPWNYPFMLAIFKVGPALAAGCTLVLKPAE 604 ADKIHG+T DGD+ ++TR EP+GV G IIPWN+P ++ +K+ PAL G T+V+KPAE Sbjct: 135 ADKIHGMTIPVDGDYFTFTRHEPIGVCGQIIPWNFPLLMFTWKIAPALCCGNTVVIKPAE 194 Query: 605 QTPLSALFLGDLILRAGFPKGVVNIVPGYGPTAGAALSSHHDVRKITFTGSTEIGHVIMK 784 QTPLSAL++G LI AGFP GVVNI+PGYGPTAGAA++SH + KI FTGSTE+G +I + Sbjct: 195 QTPLSALYMGALIKEAGFPPGVVNILPGYGPTAGAAIASHIGIDKIAFTGSTEVGKLIQE 254 Query: 785 TIGETSLKRVTLELGGKSPLIIFXXXXXXXXXXXXXXXCMTNHGQCCVAGTRTFVEASIY 964 G ++LKRVTLELGGKSP IIF N GQCC AG+R FVE SIY Sbjct: 255 AAGRSNLKRVTLELGGKSPNIIFADADLDYAVEQAHQGVFFNQGQCCTAGSRIFVEESIY 314 Query: 965 DTIIEKLKKLAEERIVGDPFDEKTIQGPQVDKEQFNKILDLIESGKHEGAKLVTGGNRIG 1144 + +++ + A+ RIVG PFD T QGPQ+DK+Q+NKIL+LI+SG EGAKL GG +G Sbjct: 315 EEFVKRSVERAKRRIVGSPFDPTTEQGPQIDKKQYNKILELIQSGVAEGAKLECGGKGLG 374 Query: 1145 NVGYFIQPTVFGDVKDNMRIAKEEIFGPVQSVFKFNTMEEVIERANNSDYGLGAGVFTSD 1324 G+FI+PTVF +V D+MRIAKEEIFGPVQ + +F TM+EVIERANNSD+GL A VFT+D Sbjct: 375 RKGFFIEPTVFSNVTDDMRIAKEEIFGPVQEILRFKTMDEVIERANNSDFGLVAAVFTND 434 Query: 1325 INKAIMMTQALEAGSVWVNCYDVGLVTAPFGGFKQSGVGRELGCYGLEPYIEVKGVTIAI 1504 INKA+M++ A++AG+VW+NCY+ +PFGGFK SG GRE+G +GL Y EVK VT+ I Sbjct: 435 INKALMVSSAMQAGTVWINCYNALNAQSPFGGFKMSGNGREMGEFGLREYSEVKTVTVKI 494 Query: 1505 PQKNS 1519 PQKNS Sbjct: 495 PQKNS 499
>sp|Q62148|AL1A2_MOUSE Retinal dehydrogenase 2 (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2) Length = 499 Score = 547 bits (1409), Expect = e-155 Identities = 270/485 (55%), Positives = 349/485 (71%), Gaps = 1/485 (0%) Frame = +2 Query: 68 EIKFTKIFINNEWCNSEEGLTFPTINPTNEQVIVQVQEGREXXXXXXXXXXXXXXXXGSE 247 EIK+TKIFINNEW NSE G FP NP + + +VQE + GS Sbjct: 15 EIKYTKIFINNEWQNSESGRVFPVCNPATGEQVCEVQEADKVDIDKAVQAARLAFSLGSV 74 Query: 248 WRTLNASARGLLLNKLADLIELEADYISRLEALDNGKSYVIAKG-DIEFSVKILRYYAGY 424 WR ++AS RG LL+KLADL+E + ++ +E+L+ GK ++ A D++ +K LRYYAG+ Sbjct: 75 WRRMDASERGRLLDKLADLVERDRATLATMESLNGGKPFLQAFYIDLQGVIKTLRYYAGW 134 Query: 425 ADKIHGVTTAPDGDFLSYTRIEPVGVVGGIIPWNYPFMLAIFKVGPALAAGCTLVLKPAE 604 ADKIHG+T DGD+ ++TR EP+GV G IIPWN+P ++ +K+ PAL G T+V+KPAE Sbjct: 135 ADKIHGMTIPVDGDYFTFTRHEPIGVCGQIIPWNFPLLMFTWKIAPALCCGNTVVIKPAE 194 Query: 605 QTPLSALFLGDLILRAGFPKGVVNIVPGYGPTAGAALSSHHDVRKITFTGSTEIGHVIMK 784 QTPLSAL++G LI AGFP GVVNI+PGYGPTAGAA++SH + KI FTGSTE+G +I + Sbjct: 195 QTPLSALYMGALIKEAGFPPGVVNILPGYGPTAGAAIASHIGIDKIAFTGSTEVGKLIQE 254 Query: 785 TIGETSLKRVTLELGGKSPLIIFXXXXXXXXXXXXXXXCMTNHGQCCVAGTRTFVEASIY 964 G ++LKRVTLELGGKSP IIF N GQCC AG+R FVE SIY Sbjct: 255 AAGRSNLKRVTLELGGKSPNIIFADADLDYAVEQAHQGVFFNQGQCCTAGSRIFVEESIY 314 Query: 965 DTIIEKLKKLAEERIVGDPFDEKTIQGPQVDKEQFNKILDLIESGKHEGAKLVTGGNRIG 1144 + +++ + A+ RIVG PFD T QGPQ+DK+Q+NK+L+LI+SG EGAKL GG +G Sbjct: 315 EEFVKRSVERAKRRIVGSPFDPTTEQGPQIDKKQYNKVLELIQSGVAEGAKLECGGKGLG 374 Query: 1145 NVGYFIQPTVFGDVKDNMRIAKEEIFGPVQSVFKFNTMEEVIERANNSDYGLGAGVFTSD 1324 G+FI+PTVF +V D+MRIAKEEIFGPVQ + +F TM+EVIERANNSD+GL A VFT+D Sbjct: 375 RKGFFIEPTVFSNVTDDMRIAKEEIFGPVQEILRFKTMDEVIERANNSDFGLVAAVFTND 434 Query: 1325 INKAIMMTQALEAGSVWVNCYDVGLVTAPFGGFKQSGVGRELGCYGLEPYIEVKGVTIAI 1504 INKA+M++ A++AG+VW+NCY+ +PFGGFK SG GRE+G +GL Y EVK VT+ I Sbjct: 435 INKALMVSSAMQAGTVWINCYNALNAQSPFGGFKMSGNGREMGEFGLREYSEVKTVTVKI 494 Query: 1505 PQKNS 1519 PQKNS Sbjct: 495 PQKNS 499
>sp|P20000|ALDH2_BOVIN Aldehyde dehydrogenase, mitochondrial precursor (ALDH class 2) (ALDHI) (ALDH-E2) Length = 520 Score = 547 bits (1409), Expect = e-155 Identities = 270/486 (55%), Positives = 343/486 (70%), Gaps = 1/486 (0%) Frame = +2 Query: 65 PEIKFTKIFINNEWCNSEEGLTFPTINPTNEQVIVQVQEGREXXXXXXXXXXXXXXXXGS 244 PE+ + +IFINNEW ++ TFPT+NP+ VI V EG + GS Sbjct: 35 PEVLYNQIFINNEWHDAVSKKTFPTVNPSTGDVICHVAEGDKADVDRAVKAARAAFQLGS 94 Query: 245 EWRTLNASARGLLLNKLADLIELEADYISRLEALDNGKSYVIAK-GDIEFSVKILRYYAG 421 WR ++AS RG LLN+LADLIE + Y++ LE LDNGK Y+I+ D++ +K LRYYAG Sbjct: 95 PWRRMDASERGRLLNRLADLIERDRTYLAALETLDNGKPYIISYLVDLDMVLKCLRYYAG 154 Query: 422 YADKIHGVTTAPDGDFLSYTRIEPVGVVGGIIPWNYPFMLAIFKVGPALAAGCTLVLKPA 601 +ADK HG T DGD+ SYTR EPVGV G IIPWN+P ++ +K+GPALA G +V+K A Sbjct: 155 WADKYHGKTIPIDGDYFSYTRHEPVGVCGQIIPWNFPLLMQAWKLGPALATGNVVVMKVA 214 Query: 602 EQTPLSALFLGDLILRAGFPKGVVNIVPGYGPTAGAALSSHHDVRKITFTGSTEIGHVIM 781 EQTPL+AL++ +LI AGFP GVVN++PG+GPTAGAA++SH DV K+ FTGSTE+GH+I Sbjct: 215 EQTPLTALYVANLIKEAGFPPGVVNVIPGFGPTAGAAIASHEDVDKVAFTGSTEVGHLIQ 274 Query: 782 KTIGETSLKRVTLELGGKSPLIIFXXXXXXXXXXXXXXXCMTNHGQCCVAGTRTFVEASI 961 G+++LKRVTLE+GGKSP II N GQCC AG+RTFV+ I Sbjct: 275 VAAGKSNLKRVTLEIGGKSPNIIMSDADMDWAVEQAHFALFFNQGQCCCAGSRTFVQEDI 334 Query: 962 YDTIIEKLKKLAEERIVGDPFDEKTIQGPQVDKEQFNKILDLIESGKHEGAKLVTGGNRI 1141 Y +E+ A+ R+VG+PFD +T QGPQVD+ QF K+L I+SGK EG KL+ GG Sbjct: 335 YAEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKVLGYIKSGKEEGLKLLCGGGAA 394 Query: 1142 GNVGYFIQPTVFGDVKDNMRIAKEEIFGPVQSVFKFNTMEEVIERANNSDYGLGAGVFTS 1321 + GYFIQPTVFGD++D M IAKEEIFGPV + KF +MEEV+ RANNS YGL A VFT Sbjct: 395 ADRGYFIQPTVFGDLQDGMTIAKEEIFGPVMQILKFKSMEEVVGRANNSKYGLAAAVFTK 454 Query: 1322 DINKAIMMTQALEAGSVWVNCYDVGLVTAPFGGFKQSGVGRELGCYGLEPYIEVKGVTIA 1501 D++KA ++QAL+AG+VWVNCYDV +PFGG+K SG GRELG YGL+ Y EVK VT+ Sbjct: 455 DLDKANYLSQALQAGTVWVNCYDVFGAQSPFGGYKLSGSGRELGEYGLQAYTEVKTVTVR 514 Query: 1502 IPQKNS 1519 +PQKNS Sbjct: 515 VPQKNS 520
>sp|O94788|AL1A2_HUMAN Retinal dehydrogenase 2 (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2) Length = 499 Score = 543 bits (1400), Expect = e-154 Identities = 268/485 (55%), Positives = 346/485 (71%), Gaps = 1/485 (0%) Frame = +2 Query: 68 EIKFTKIFINNEWCNSEEGLTFPTINPTNEQVIVQVQEGREXXXXXXXXXXXXXXXXGSE 247 EIK+TKIFINNEW NSE G FP NP + + +VQE + GS Sbjct: 15 EIKYTKIFINNEWQNSESGRVFPVYNPATGEQVCEVQEADKADIDKAVQAARLAFSLGSV 74 Query: 248 WRTLNASARGLLLNKLADLIELEADYISRLEALDNGKSYVIA-KGDIEFSVKILRYYAGY 424 WR ++AS RG LL+KLADL+E + ++ +E+L+ GK ++ A D++ +K RYYAG+ Sbjct: 75 WRRMDASERGRLLDKLADLVERDRAVLATMESLNGGKPFLQAFYVDLQGVIKTFRYYAGW 134 Query: 425 ADKIHGVTTAPDGDFLSYTRIEPVGVVGGIIPWNYPFMLAIFKVGPALAAGCTLVLKPAE 604 ADKIHG+T DGD+ ++TR EP+GV G IIPWN+P ++ +K+ PAL G T+V+KPAE Sbjct: 135 ADKIHGMTIPVDGDYFTFTRHEPIGVCGQIIPWNFPLLMFAWKIAPALCCGNTVVIKPAE 194 Query: 605 QTPLSALFLGDLILRAGFPKGVVNIVPGYGPTAGAALSSHHDVRKITFTGSTEIGHVIMK 784 QTPLSAL++G LI AGFP GV+NI+PGYGPTAGAA++SH + KI FTGSTE+G +I + Sbjct: 195 QTPLSALYMGALIKEAGFPPGVINILPGYGPTAGAAIASHIGIDKIAFTGSTEVGKLIQE 254 Query: 785 TIGETSLKRVTLELGGKSPLIIFXXXXXXXXXXXXXXXCMTNHGQCCVAGTRTFVEASIY 964 G ++LKRVTLELGGKSP IIF N GQCC AG+R FVE SIY Sbjct: 255 AAGRSNLKRVTLELGGKSPNIIFADADLDYAVEQAHQGVFFNQGQCCTAGSRIFVEESIY 314 Query: 965 DTIIEKLKKLAEERIVGDPFDEKTIQGPQVDKEQFNKILDLIESGKHEGAKLVTGGNRIG 1144 + + + + A+ RIVG PFD T QGPQ+DK+Q+NKIL+LI+SG EGAKL GG +G Sbjct: 315 EEFVRRSVERAKRRIVGSPFDPTTEQGPQIDKKQYNKILELIQSGVAEGAKLECGGKGLG 374 Query: 1145 NVGYFIQPTVFGDVKDNMRIAKEEIFGPVQSVFKFNTMEEVIERANNSDYGLGAGVFTSD 1324 G+FI+PTVF +V D+MRIAKEEIFGPVQ + +F TM+EVIERANNSD+GL A VFT+D Sbjct: 375 RKGFFIEPTVFSNVTDDMRIAKEEIFGPVQEILRFKTMDEVIERANNSDFGLVAAVFTND 434 Query: 1325 INKAIMMTQALEAGSVWVNCYDVGLVTAPFGGFKQSGVGRELGCYGLEPYIEVKGVTIAI 1504 INKA+ ++ A++AG+VW+NCY+ +PFGGFK SG GRE+G +GL Y EVK VT+ I Sbjct: 435 INKALTVSSAMQAGTVWINCYNALNAQSPFGGFKMSGNGREMGEFGLREYSEVKTVTVKI 494 Query: 1505 PQKNS 1519 PQKNS Sbjct: 495 PQKNS 499
>sp|P27463|AL1A1_CHICK Retinal dehydrogenase 1 (RalDH1) (RALDH 1) (Aldehyde dehydrogenase family 1 member A1) (Aldehyde dehydrogenase, cytosolic) (ALHDII) (ALDH-E1) Length = 509 Score = 540 bits (1392), Expect = e-153 Identities = 276/485 (56%), Positives = 339/485 (69%), Gaps = 1/485 (0%) Frame = +2 Query: 68 EIKFTKIFINNEWCNSEEGLTFPTINPTNEQVIVQVQEGREXXXXXXXXXXXXXXXXGSE 247 +IK+TKIFINNEW +S G F NP NE+ I +V EG + GS Sbjct: 25 KIKYTKIFINNEWHDSVSGKKFEVFNPANEEKICEVAEGDKADIDKAVKAARKAFELGSP 84 Query: 248 WRTLNASARGLLLNKLADLIELEADYISRLEALDNGKSYVIAK-GDIEFSVKILRYYAGY 424 WRT++AS RG LLNKLADL+E + ++ +EA+D GK + A D+ +K +RY AG+ Sbjct: 85 WRTMDASERGRLLNKLADLVERDRLTLATMEAIDGGKLFSTAYLMDLGACIKTIRYCAGW 144 Query: 425 ADKIHGVTTAPDGDFLSYTRIEPVGVVGGIIPWNYPFMLAIFKVGPALAAGCTLVLKPAE 604 ADKIHG T DG+F ++TR EPVGV G IIPWN+P ++ I+K+ PAL G T+V+KPAE Sbjct: 145 ADKIHGRTVPMDGNFFTFTRHEPVGVCGQIIPWNFPLVMFIWKIAPALCCGNTVVVKPAE 204 Query: 605 QTPLSALFLGDLILRAGFPKGVVNIVPGYGPTAGAALSSHHDVRKITFTGSTEIGHVIMK 784 QTPLSAL++G LI AGFP GVVNIVPG+GPTAGAA+S H D+ K++FTGSTE+G +I + Sbjct: 205 QTPLSALYMGSLIKEAGFPPGVVNIVPGFGPTAGAAISHHMDIDKVSFTGSTEVGKLIKE 264 Query: 785 TIGETSLKRVTLELGGKSPLIIFXXXXXXXXXXXXXXXCMTNHGQCCVAGTRTFVEASIY 964 G+T+LKRVTLELGGKSP IIF + GQCC+AG+R FVE IY Sbjct: 265 AAGKTNLKRVTLELGGKSPNIIFADADLDEAAEFAHIGLFYHQGQCCIAGSRIFVEEPIY 324 Query: 965 DTIIEKLKKLAEERIVGDPFDEKTIQGPQVDKEQFNKILDLIESGKHEGAKLVTGGNRIG 1144 D + + + A++ +GDP QGPQ+DKEQF KILDLIESGK EGAKL GG G Sbjct: 325 DEFVRRSIERAKKYTLGDPLLPGVQQGPQIDKEQFQKILDLIESGKKEGAKLECGGGPWG 384 Query: 1145 NVGYFIQPTVFGDVKDNMRIAKEEIFGPVQSVFKFNTMEEVIERANNSDYGLGAGVFTSD 1324 N GYFIQPTVF +V D+MRIAKEEIFGPVQ + KF T++EVI+RANN+ YGL A VFT D Sbjct: 385 NKGYFIQPTVFSNVTDDMRIAKEEIFGPVQQIMKFKTIDEVIKRANNTTYGLAAAVFTKD 444 Query: 1325 INKAIMMTQALEAGSVWVNCYDVGLVTAPFGGFKQSGVGRELGCYGLEPYIEVKGVTIAI 1504 I+KA+ AL+AG+VWVNCY PFGGFK SG GRELG YGL+ Y EVK VTI I Sbjct: 445 IDKALTFASALQAGTVWVNCYSAFSAQCPFGGFKMSGNGRELGEYGLQEYTEVKTVTIKI 504 Query: 1505 PQKNS 1519 PQKNS Sbjct: 505 PQKNS 509
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 165,657,367
Number of Sequences: 369166
Number of extensions: 3395105
Number of successful extensions: 9569
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8401
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8913
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 19503728370
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail