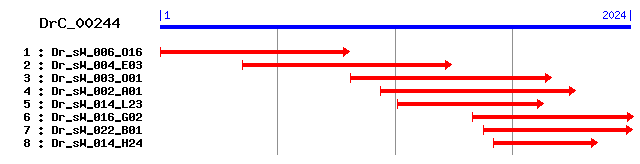
DrC_00244
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00244 (2020 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P90901|IFA1_CAEEL Intermediate filament protein ifa-1 (I... 236 2e-61 sp|Q01241|NF70_LOLPE 70 kDa neurofilament protein (NF70) 235 4e-61 sp|Q01240|NF60_LOLPE 60 kDa neurofilament protein (NF60) 235 4e-61 sp|O02365|IFA2_CAEEL Intermediate filament protein ifa-2 (I... 224 7e-58 sp|P90900|IFA4_CAEEL Intermediate filament protein ifa-4 (I... 223 1e-57 sp|P23731|IFEB_ASCSU Intermediate filament protein B (IF-B) 205 3e-52 sp|Q21065|IFA3_CAEEL Intermediate filament protein ifa-3 (I... 204 1e-51 sp|P23730|IFEA_ASCSU Intermediate filament protein A (IF-A) 202 4e-51 sp|P31732|OV71_ONCVO Muscle cell intermediate filament prot... 200 1e-50 sp|Q19289|IFB1_CAEEL Intermediate filament protein ifb-1 (I... 190 1e-47
>sp|P90901|IFA1_CAEEL Intermediate filament protein ifa-1 (Intermediate filament protein A1) (IF-A1) (Cel IF A1) Length = 575 Score = 236 bits (601), Expect = 2e-61 Identities = 152/512 (29%), Positives = 254/512 (49%), Gaps = 3/512 (0%) Frame = +1 Query: 403 SCEIEQLKGGSVKDTSKIKEIYEVEIRQLRNLLNDSEKNRAPLEVQIDSLEYELDDLRKR 582 + +++ L+ KDT I+ +YE E+ + L++++ K R +E Q+ ++ EL ++R++ Sbjct: 98 AADLDALRSKWGKDTHNIRNMYEGELVDAQKLIDETNKQRKDMEGQLKKMQDELAEMRRK 157 Query: 583 LMLLEDINGQYKSTIDKQRQELGNFEGELVELRRFKNTRNADVSKEKEEVKSLRDELKKL 762 L Q ++ ID L N E E+ L+R +V + K+E + L EL++ Sbjct: 158 LEDATKGREQDRAKIDALLVTLSNLEAEISLLKRRIAQLEDEVKRIKQENQRLLSELQRA 217 Query: 763 RQDYEREALAHITVEHELQNLRETMDFEKQIHETEVKELTTVAYRDPTKENMMYWQSEMG 942 R D ++E L I ++++Q L E +DF +++H+ E+KEL T+A RD T EN ++++E+ Sbjct: 218 RTDLDQETLNRIDYQNQVQTLLEEIDFLRRVHDNEIKELQTLASRDTTPENREFFKNELS 277 Query: 943 RALRDIQTEYENKLSGIKQEMEASYGLQMQEIRTSKTRDNVETTHYKEENSRIKSQIQSM 1122 A+RDI+ EY+ + + +ME+ Y L++QEI+T R N+E + KEE R+++Q+ + Sbjct: 278 SAIRDIREEYDQVNNVHRNDMESWYRLKVQEIQTQSARQNMEQGYAKEEVKRLRTQLSDL 337 Query: 1123 RSRIADLEMQNAEYEHNLNMIRGNYEETQRSLELKLMSXXXXXXXXXXXXSAALTELQSL 1302 R ++ADLE +N+ E + + E+ QRS E L A + ELQ L Sbjct: 338 RGKLADLESRNSLLEKQIQELNYQLEDDQRSYEAALNDRDSQIRKMREECQALMVELQML 397 Query: 1303 LDAKLGLELEIAAYAKLLEFEESRVNSNTEGNMIISQFXXXXXXXXXXXXXXXXXXXXXX 1482 LD K L+ EIA Y K+LE EE+R ++ Sbjct: 398 LDTKQTLDAEIAIYRKMLEGEENRAGLKQLVEQVVKTHAITQETDTE------------- 444 Query: 1483 XXXXXXXXXXXXXXXXXXXXXMTERETTDYQSKRMKFRRTTKGNISIAETDLTGSFIILE 1662 T R + R F+R+ KGN+SI E G FI+L+ Sbjct: 445 ----------------------TMRVVKGETASRQSFQRSAKGNVSIHEASPDGKFIVLQ 482 Query: 1663 NI-GTKVESLEGMTIKRAIEGNSNQISYTISSKVTLNAGEKISLVSRGKKTGKGLE--IE 1833 N K E++ +KR I+G + YT+ L AG+ + + +R + + + Sbjct: 483 NTHRAKDEAIGEWKLKRRIDGKRENV-YTLPRDFVLRAGKTLKIFARNQGVASPPDQLVY 541 Query: 1834 TDEASWGVGNNVVTTSYDSEGVEKAAHYQEVS 1929 E S+G GNNV T ++ EG E+A H Q S Sbjct: 542 DAEDSFGSGNNVQTILFNKEGEERATHIQRQS 573
Score = 54.3 bits (129), Expect = 1e-06
Identities = 24/42 (57%), Positives = 34/42 (80%)
Frame = +2
Query: 275 QSNVNSMRKGRAQEKRDMQDLNERFASYIEKARFLEAQNKQL 400
Q+ +++R R +EK++M DLN+R ASYIEK RFLEAQN++L
Sbjct: 56 QNAASTIRDSREREKKEMSDLNDRLASYIEKVRFLEAQNRKL 97
>sp|Q01241|NF70_LOLPE 70 kDa neurofilament protein (NF70) Length = 615 Score = 235 bits (599), Expect = 4e-61 Identities = 133/369 (36%), Positives = 212/369 (57%), Gaps = 2/369 (0%) Frame = +1 Query: 277 IKCKLDEERKSPR--KERYARFE*KIRQLY*KGTIPRSSK*TISSCEIEQLKGGSVKDTS 450 +K + E++ R ER+A + K+R L ++ + E+E+LK K+TS Sbjct: 89 VKANREREKQDMRDLNERFANYIEKVRFL--------EAQNKKLAGELEELKSKWGKETS 140 Query: 451 KIKEIYEVEIRQLRNLLNDSEKNRAPLEVQIDSLEYELDDLRKRLMLLEDINGQYKSTID 630 IKE+YE E+ + R L++ + K + L+V++ L +L+ +K L + + I Sbjct: 141 AIKEMYETELEEARKLIDATNKEKITLDVRVTELIDQLERQQKDLEESRTYHQIDQEQIA 200 Query: 631 KQRQELGNFEGELVELRRFKNTRNADVSKEKEEVKSLRDELKKLRQDYEREALAHITVEH 810 +Q Q+L + EGE+ LRR + + ++ + + DE++K+R D E + H+ E+ Sbjct: 201 RQNQQLADLEGEISMLRRSIESLEKEKMRQSNILAKMNDEMEKMRMDLNNETINHLDAEN 260 Query: 811 ELQNLRETMDFEKQIHETEVKELTTVAYRDPTKENMMYWQSEMGRALRDIQTEYENKLSG 990 Q L E ++F+K +H E+KEL +AYRD T EN +W++E+ +A+RDIQ EY+ K Sbjct: 261 RRQTLEEELEFQKDVHAQELKELAALAYRDTTAENREFWRNELAQAIRDIQQEYDAKCDQ 320 Query: 991 IKQEMEASYGLQMQEIRTSKTRDNVETTHYKEENSRIKSQIQSMRSRIADLEMQNAEYEH 1170 ++ ++EA Y L++QE RT T+ N+E T KEEN+++KS + +R+R+ADLE +NA+ E Sbjct: 321 MRGDIEAYYNLKVQEFRTGATKQNMEVTRNKEENTKLKSNMTEIRNRLADLEARNAQLER 380 Query: 1171 NLNMIRGNYEETQRSLELKLMSXXXXXXXXXXXXSAALTELQSLLDAKLGLELEIAAYAK 1350 + + EE R EL+ + L ELQ L+D KL LELEIAAY K Sbjct: 381 TNQDLLRDLEEKDRQNELESCQYKEEITKLRGEMESILKELQDLMDIKLSLELEIAAYRK 440 Query: 1351 LLEFEESRV 1377 LLE EESRV Sbjct: 441 LLEGEESRV 449
Score = 59.7 bits (143), Expect = 3e-08
Identities = 27/41 (65%), Positives = 35/41 (85%)
Frame = +2
Query: 278 SNVNSMRKGRAQEKRDMQDLNERFASYIEKARFLEAQNKQL 400
+ VN ++ R +EK+DM+DLNERFA+YIEK RFLEAQNK+L
Sbjct: 84 TGVNHVKANREREKQDMRDLNERFANYIEKVRFLEAQNKKL 124
Score = 55.1 bits (131), Expect = 8e-07
Identities = 39/113 (34%), Positives = 61/113 (53%), Gaps = 3/113 (2%)
Frame = +1
Query: 1591 FRRTTKGNISIAETDLTGSFIILENIGTKVESLEGMTIKRAIEGNSNQISYTISSKVTLN 1770
++RT+KG++SI E D G FI LE K E+L G I R ++ N +Y I + V L
Sbjct: 503 YQRTSKGSVSIKEADSQGCFIALET--KKEENLTGWKIVRKVDDNK-VYTYEIPNLV-LK 558
Query: 1771 AGEKISLVSRGKKT---GKGLEIETDEASWGVGNNVVTTSYDSEGVEKAAHYQ 1920
G + + S+ + G L + + +WG G+NVVT + +G +KA + Q
Sbjct: 559 TGTVVKIWSKNHQAQARGDDL-VSRENDTWGTGSNVVTILQNEKGEDKANYTQ 610
>sp|Q01240|NF60_LOLPE 60 kDa neurofilament protein (NF60) Length = 511 Score = 235 bits (599), Expect = 4e-61 Identities = 133/369 (36%), Positives = 212/369 (57%), Gaps = 2/369 (0%) Frame = +1 Query: 277 IKCKLDEERKSPR--KERYARFE*KIRQLY*KGTIPRSSK*TISSCEIEQLKGGSVKDTS 450 +K + E++ R ER+A + K+R L ++ + E+E+LK K+TS Sbjct: 89 VKANREREKQDMRDLNERFANYIEKVRFL--------EAQNKKLAGELEELKSKWGKETS 140 Query: 451 KIKEIYEVEIRQLRNLLNDSEKNRAPLEVQIDSLEYELDDLRKRLMLLEDINGQYKSTID 630 IKE+YE E+ + R L++ + K + L+V++ L +L+ +K L + + I Sbjct: 141 AIKEMYETELEEARKLIDATNKEKITLDVRVTELIDQLERQQKDLEESRTYHQIDQEQIA 200 Query: 631 KQRQELGNFEGELVELRRFKNTRNADVSKEKEEVKSLRDELKKLRQDYEREALAHITVEH 810 +Q Q+L + EGE+ LRR + + ++ + + DE++K+R D E + H+ E+ Sbjct: 201 RQNQQLADLEGEISMLRRSIESLEKEKMRQSNILAKMNDEMEKMRMDLNNETINHLDAEN 260 Query: 811 ELQNLRETMDFEKQIHETEVKELTTVAYRDPTKENMMYWQSEMGRALRDIQTEYENKLSG 990 Q L E ++F+K +H E+KEL +AYRD T EN +W++E+ +A+RDIQ EY+ K Sbjct: 261 RRQTLEEELEFQKDVHAQELKELAALAYRDTTAENREFWRNELAQAIRDIQQEYDAKCDQ 320 Query: 991 IKQEMEASYGLQMQEIRTSKTRDNVETTHYKEENSRIKSQIQSMRSRIADLEMQNAEYEH 1170 ++ ++EA Y L++QE RT T+ N+E T KEEN+++KS + +R+R+ADLE +NA+ E Sbjct: 321 MRGDIEAYYNLKVQEFRTGATKQNMEVTRNKEENTKLKSNMTEIRNRLADLEARNAQLER 380 Query: 1171 NLNMIRGNYEETQRSLELKLMSXXXXXXXXXXXXSAALTELQSLLDAKLGLELEIAAYAK 1350 + + EE R EL+ + L ELQ L+D KL LELEIAAY K Sbjct: 381 TNQDLLRDLEEKDRQNELESCQYKEEITKLRGEMESILKELQDLMDIKLSLELEIAAYRK 440 Query: 1351 LLEFEESRV 1377 LLE EESRV Sbjct: 441 LLEGEESRV 449
Score = 59.7 bits (143), Expect = 3e-08
Identities = 27/41 (65%), Positives = 35/41 (85%)
Frame = +2
Query: 278 SNVNSMRKGRAQEKRDMQDLNERFASYIEKARFLEAQNKQL 400
+ VN ++ R +EK+DM+DLNERFA+YIEK RFLEAQNK+L
Sbjct: 84 TGVNHVKANREREKQDMRDLNERFANYIEKVRFLEAQNKKL 124
>sp|O02365|IFA2_CAEEL Intermediate filament protein ifa-2 (Intermediate filament protein A2) (IF-A2) (Cel IF A2) Length = 581 Score = 224 bits (571), Expect = 7e-58 Identities = 154/511 (30%), Positives = 250/511 (48%), Gaps = 4/511 (0%) Frame = +1 Query: 409 EIEQLKGGSVKDTSKIKEIYEVEIRQLRNLLNDSEKNRAPLEVQIDSLEYELDDLRKRLM 588 +++ L+G K T +K +YE+EI N++ + K+ E +I L+ +LD+LRK+ Sbjct: 102 DLKMLQGRFGKSTGSVKVMYEMEITTATNVVKQTGKDHGETEKEIRKLQDQLDELRKKFE 161 Query: 589 LLEDINGQYKSTIDKQRQELGNFEGELVELRRFKNTRNADVSKEKEEVKSLRDELKKLRQ 768 + + + ID L N E E+ L+R +V++ K+E L EL+++R Sbjct: 162 EAQRGRAEDRLKIDDLLVTLSNLEAEINLLKRRIALLEEEVARLKKENFRLTSELQRVRI 221 Query: 769 DYEREALAHITVEHELQNLRETMDFEKQIHETEVKELTTVAYRDPTKENMMYWQSEMGRA 948 + ++E L I +++++ + E +DF K+ ETE+KEL A RD T EN Y+++E+ A Sbjct: 222 ELDQETLLRIDNQNKVKTILEEIDFMKRGFETELKELQAQAARDTTSENREYFKNELANA 281 Query: 949 LRDIQTEYENKLSGIKQEMEASYGLQMQEIRTSKTRDNVETTHYKEENSRIKSQIQSMRS 1128 +RDI+ EY+ ++G + +ME+ Y L++QEI T R N E + KEE R+++Q +R Sbjct: 282 MRDIRAEYDQIMNGNRNDMESWYQLRVQEINTQSNRQNAENNYQKEEVKRLRNQTSELRQ 341 Query: 1129 RIADLEMQNAEYEHNLNMIRGNYEETQRSLELKLMSXXXXXXXXXXXXSAALTELQSLLD 1308 +++DLE +N E + + E+ QRS E L A + ELQ LLD Sbjct: 342 KLSDLESRNLLLEKQIEDLNYQLEDDQRSYEAALNDKDAQIRKLREECQALMVELQMLLD 401 Query: 1309 AKLGLELEIAAYAKLLEFEESRVNSNTEGNMIISQFXXXXXXXXXXXXXXXXXXXXXXXX 1488 K L+ E+ Y ++LE N+EGN + Sbjct: 402 TKQTLDGELKVYRQMLE-------GNSEGNGL---------------------------- 426 Query: 1489 XXXXXXXXXXXXXXXXXXXMTERETTDYQSKRMKFRRTTKGNISIAETDLTGSFIILENI 1668 T R S R ++R+ KGN++I ET G F+ILEN Sbjct: 427 RQLVEKVVRTSAINEEADTETMRVVKGEHSSRTSYQRSAKGNVAIKETSPEGKFVILENT 486 Query: 1669 -GTKVESLEGMTIKRAIEGNSNQISYTISSKVTLNAGEKISLVSRGKKTGKGLEI---ET 1836 K E L +KR I+G +I +T S L+ + + + +RG+ E+ E Sbjct: 487 HRAKEEPLGDWKLKRKIDG-KREIVFTFPSDYILHPFQSVKIFARGQGIANPPEVLIFEG 545 Query: 1837 DEASWGVGNNVVTTSYDSEGVEKAAHYQEVS 1929 DE ++GVG NV T Y+++G E+A H Q S Sbjct: 546 DE-TFGVGANVQTILYNNKGEERATHIQRQS 575
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/42 (50%), Positives = 34/42 (80%)
Frame = +2
Query: 275 QSNVNSMRKGRAQEKRDMQDLNERFASYIEKARFLEAQNKQL 400
Q+ +++R R +EK+++ +LN+R ASYIEK RFL+AQN++L
Sbjct: 58 QNAASTIRDNREREKKEIMELNDRLASYIEKVRFLDAQNRKL 99
>sp|P90900|IFA4_CAEEL Intermediate filament protein ifa-4 (Intermediate filament protein A4) (IF-A4) (Cel IF A4) Length = 575 Score = 223 bits (569), Expect = 1e-57 Identities = 149/513 (29%), Positives = 252/513 (49%), Gaps = 6/513 (1%) Frame = +1 Query: 409 EIEQLKGGSVKDTSKIKEIYEVEIRQLRNLLNDSEKNRAPLEVQIDSLEYELDDLRKRLM 588 ++ L+G D++ +K +YE E+R ++L+ DS+K RA LE QI L EL++ R +L Sbjct: 101 DLNLLRGKWGHDSTSVKVMYETELRSAKDLIADSDKERAQLEDQIKKLVEELNNYRNKLH 160 Query: 589 LLEDINGQYKSTIDKQRQELGNFEGELVELRRFKNTRNADVSKEKEEVKSLRDELKKLRQ 768 E + +D LG E E+ L+R + + K E + D L R Sbjct: 161 EAERGADVTRKELDNVLGRLGALEAEIELLKRRISLMEESIGHLKRENHRMIDSLHHARN 220 Query: 769 DYEREALAHITVEHELQNLRETMDFEKQIHETEVKELTTVAYRDPTKENMMYWQSEMGRA 948 E+E L I ++++Q L E DF ++IH++E+ +L +A RD T EN Y+++E+ A Sbjct: 221 AVEQETLNRIDYQNQVQTLLEECDFIRRIHDSEIHDLLAMASRDTTVENREYFKNELSSA 280 Query: 949 LRDIQTEYENKLSGIKQEMEASYGLQMQEIRTSKTRDNVETTHYKEENSRIKSQIQSMRS 1128 +R+I++EY++ + + ++E+ Y L++QEI T +R+ +E + +EE R+++ + MR Sbjct: 281 IREIRSEYDHVSNTTRTDIESWYKLKVQEIHTQNSRNCLEQNYAREEVKRLRTTLGDMRG 340 Query: 1129 RIADLEMQNAEYEHNLNMIRGNYEETQRSLELKLMSXXXXXXXXXXXXSAALTELQSLLD 1308 ++ADLE +N E + + EE RS E L + ELQ L+D Sbjct: 341 KMADLEGRNLLLEKQIEDLNYQMEEDMRSYEQSLNDKDTSINKLRDESKILMVELQMLID 400 Query: 1309 AKLGLELEIAAYAKLLEFEESRVNSNTEGNMIISQFXXXXXXXXXXXXXXXXXXXXXXXX 1488 K L+ EI Y K+L+ EE+R ++ Sbjct: 401 TKQTLDAEIVIYRKMLDGEENRAGLRQLVEQVVK-------------------------- 434 Query: 1489 XXXXXXXXXXXXXXXXXXXMTERETTDYQSKRMKFRRTTKGNISIAETDLTGSFIILENI 1668 + + ETT + S + R+ KGNI+I E + +G +I+LENI Sbjct: 435 -----TTSIHQTKEIESLRVLKGETTSHSS----YSRSAKGNIAIQEAEPSGKYIVLENI 485 Query: 1669 GTKVESLEGMTIKRAIEGNSNQISYTISSKVTLNAGEKISLVSRGKKTGKGLEIETD--- 1839 + E++ ++R I G +I +T + TL A + + + +R G+G+ D Sbjct: 486 SRRDENIGDWKLRRKIAG-KREIVFTFpreFTLRAQKSVKIFAR----GQGVHSPPDSLV 540 Query: 1840 ---EASWGVGNNVVTTSYDSEGVEKAAHYQEVS 1929 E S+G GN+VVTT Y+ EG ++A+H Q S Sbjct: 541 YDLEDSFGTGNDVVTTLYNKEGEDRASHSQRAS 573
Score = 51.6 bits (122), Expect = 8e-06
Identities = 21/38 (55%), Positives = 33/38 (86%)
Frame = +2
Query: 287 NSMRKGRAQEKRDMQDLNERFASYIEKARFLEAQNKQL 400
+++R+ R +EK++M +LN+R ASYIEK RFLEAQN+++
Sbjct: 61 SAIRESRTREKKEMSELNDRLASYIEKVRFLEAQNRKM 98
Score = 36.2 bits (82), Expect = 0.37
Identities = 25/80 (31%), Positives = 40/80 (50%)
Frame = +1
Query: 985 SGIKQEMEASYGLQMQEIRTSKTRDNVETTHYKEENSRIKSQIQSMRSRIADLEMQNAEY 1164
S + M G IR S+TR+ E + E N R+ S I+ +R LE QN +
Sbjct: 46 SNLTSGMSPFGGHAASAIRESRTREKKEMS---ELNDRLASYIEKVRF----LEAQNRKM 98
Query: 1165 EHNLNMIRGNYEETQRSLEL 1224
E +LN++RG + S+++
Sbjct: 99 EKDLNLLRGKWGHDSTSVKV 118
>sp|P23731|IFEB_ASCSU Intermediate filament protein B (IF-B) Length = 589 Score = 205 bits (522), Expect = 3e-52 Identities = 141/509 (27%), Positives = 244/509 (47%), Gaps = 4/509 (0%) Frame = +1 Query: 409 EIEQLKGGSVKDTSKIKEIYEVEIRQLRNLLNDSEKNRAPLEVQIDSLEYELDDLRKRLM 588 ++++L+G KDTS+IK Y +R R ++D + +A ++V++ L +L +LR R Sbjct: 112 DLDELRGRWGKDTSEIKIQYSDSLRDARKEIDDGARRKAEIDVKVARLRDDLAELRNRYE 171 Query: 589 LLEDINGQYKSTIDKQRQELGNFEGELVELRRFKNTRNADVSKEKEEVKSLRDELKKLRQ 768 ++ + I++ + + + + EL LR + + + + +EL+K R Sbjct: 172 DVQHRRESDREKINQWQHAIEDAQSELEMLRARWRQLTEEEKRLNGDNARIWEELQKARN 231 Query: 769 DYEREALAHITVEHELQNLRETMDFEKQIHETEVKELTTVAYRDPTKENMMYWQSEMGRA 948 D + E L I ++++Q L E ++F +++HE EVKEL + + P + ++++E+ A Sbjct: 232 DLDEETLGRIDFQNQVQTLMEELEFLRRVHEQEVKELQALLAQAPA-DTREFFKNELALA 290 Query: 949 LRDIQTEYENKLSGIKQEMEASYGLQMQEIRTSKTRDNVETTHYKEENSRIKSQIQSMRS 1128 +RDI+ EY+ KQ+ME+ Y L++ E++ S R N+E+++ +EE R++ I +R Sbjct: 291 IRDIKDEYDYIAKQGKQDMESWYKLKVSEVQGSANRANMESSYQREEVKRMRDNIGDLRG 350 Query: 1129 RIADLEMQNAEYEHNLNMIRGNYEETQRSLELKLMSXXXXXXXXXXXXSAALTELQSLLD 1308 ++ DLE +NA E + + + QR E L + ELQ+LLD Sbjct: 351 KLGDLEAKNALLEKEVQNLNYQLNDDQRQYEAALNDRDATLRRMREECQTLVAELQALLD 410 Query: 1309 AKLGLELEIAAYAKLLEFEESRVNSNTEGNMIISQFXXXXXXXXXXXXXXXXXXXXXXXX 1488 K L+ EIA Y K+LE EESRV ++ Sbjct: 411 TKQMLDAEIAIYRKMLEGEESRVGLRQMVEQVVKTHSLQQQEDTD--------------- 455 Query: 1489 XXXXXXXXXXXXXXXXXXXMTERETTDYQSKRMKFRRTTKGNISIAETDLTGSFIILENI 1668 + R S + F+R+ KGN++I+E D G FI LEN Sbjct: 456 --------------------STRNVRGEVSTKTTFQRSAKGNVTISECDPNGKFITLENT 495 Query: 1669 -GTKVESLEGMTIKRAIEGNSNQISYTISSKVTLNAGEKISLVSR---GKKTGKGLEIET 1836 +K E+L +KR ++ N +I YTI L AG + + +R G + Sbjct: 496 HRSKDENLGEHRLKRKLD-NRREIVYTIPPNTVLKAGRTMKIYARDQGGIHNPPDTLVFD 554 Query: 1837 DEASWGVGNNVVTTSYDSEGVEKAAHYQE 1923 E +WG+G NVVT+ + +G E+A H Q+ Sbjct: 555 GENTWGIGANVVTSLINKDGDERATHTQK 583
Score = 45.4 bits (106), Expect = 6e-04
Identities = 30/97 (30%), Positives = 51/97 (52%), Gaps = 9/97 (9%)
Frame = +2
Query: 140 YKPISQPRNNLRDLQKTAGISLNAQMSDQQGGSMVPLIR--------GDSVALQSNV-NS 292
Y+ QPR +R + +G +S GG ++ ++ G S AL +N S
Sbjct: 15 YRSTIQPRTAVRTQSRQSGAYSTGAVSGG-GGRVLKMVTEMGSASIGGISPALSANAAKS 73
Query: 293 MRKGRAQEKRDMQDLNERFASYIEKARFLEAQNKQLV 403
+ +EK++MQ LN+R +YI++ + LE QN++LV
Sbjct: 74 FLEATDKEKKEMQGLNDRLGNYIDRVKKLEEQNRKLV 110
>sp|Q21065|IFA3_CAEEL Intermediate filament protein ifa-3 (Intermediate filament protein A3) (IF-A3) (Cel IF A3) Length = 581 Score = 204 bits (518), Expect = 1e-51 Identities = 143/500 (28%), Positives = 236/500 (47%), Gaps = 3/500 (0%) Frame = +1 Query: 439 KDTSKIKEIYEVEIRQLRNLLNDSEKNRAPLEVQIDSLEYELDDLRKRLMLLEDINGQYK 618 K T +K +YE+EI N++ ++ K+ E +I ++ +LD+LRK+ + + + Sbjct: 112 KSTGSVKIMYEMEITTATNVVKETGKDHEEAEKEIGKIKDQLDELRKKFEEAQKGRAEDR 171 Query: 619 STIDKQRQELGNFEGELVELRRFKNTRNADVSKEKEEVKSLRDELKKLRQDYEREALAHI 798 ID+ L N E E+ L+R +V++ K+E L EL+++R + ++E L I Sbjct: 172 LKIDELLVTLSNLEAEINLLKRRIALLEEEVARLKKENFRLTSELQRVRSELDQETLLRI 231 Query: 799 TVEHELQNLRETMDFEKQIHETEVKELTTVAYRDPTKENMMYWQSEMGRALRDIQTEYEN 978 ++++ + E +DF K+ ETE+K+L A RD T EN Y+++E+ ++RDI+ EY+ Sbjct: 232 DNQNKVTTILEEIDFMKRGFETELKDLQAQAARDTTSENREYFKNELMNSIRDIRAEYDR 291 Query: 979 KLSGIKQEMEASYGLQMQEIRTSKTRDNVETTHYKEENSRIKSQIQSMRSRIADLEMQNA 1158 ++G + ++E+ +++QEI T R N E H ++E R+ SQ+ ++S+ A+L +N Sbjct: 292 FMAGNRNDLESWSQIRVQEINTQTNRQNAEINHKRDEVKRLHSQVSELKSKHAELAARNG 351 Query: 1159 EYEHNLNMIRGNYEETQRSLELKLMSXXXXXXXXXXXXSAALTELQSLLDAKLGLELEIA 1338 E L + E+ QRS E L A L ELQ LLD K L+ E+ Sbjct: 352 LLEKQLEDLNYQLEDDQRSYEAALNDKDAQVRKLREECQALLVELQMLLDTKQTLDGELK 411 Query: 1339 AYAKLLEFEESRVNSNTEGNMIISQFXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX 1518 Y ++LE N+E N + Sbjct: 412 VYRRMLE-------GNSEENGL----------------------------RQLVEKVVRT 436 Query: 1519 XXXXXXXXXMTERETTDYQSKRMKFRRTTKGNISIAETDLTGSFIILENI-GTKVESLEG 1695 T R S R ++R+ KGN+SI E G F+ILEN K E L Sbjct: 437 SAINEEVDTETMRVVKGEHSSRTSYQRSAKGNVSIKEVSPEGKFVILENTHRDKEEPLGD 496 Query: 1696 MTIKRAIEGNSNQISYTISSKVTLNAGEKISLVSRGKKTGKGLEIETDEA--SWGVGNNV 1869 +KR I+G +I +T S L+ + + + +RG E+ E ++G G NV Sbjct: 497 WKLKRKIDG-KREIVFTFPSDYILHPVQTVKIFARGNGVANPPEVLVFEGDDTFGAGANV 555 Query: 1870 VTTSYDSEGVEKAAHYQEVS 1929 T Y++ G E+A H Q S Sbjct: 556 QTILYNNSGEERATHMQRQS 575
Score = 45.1 bits (105), Expect = 8e-04
Identities = 20/42 (47%), Positives = 31/42 (73%)
Frame = +2
Query: 275 QSNVNSMRKGRAQEKRDMQDLNERFASYIEKARFLEAQNKQL 400
Q +++R R +EK+++ +LN+R ASYI K RFL AQN++L
Sbjct: 58 QGAASTIRDDREREKKEITELNDRLASYIGKVRFLAAQNRKL 99
>sp|P23730|IFEA_ASCSU Intermediate filament protein A (IF-A) Length = 497 Score = 202 bits (513), Expect = 4e-51 Identities = 151/520 (29%), Positives = 255/520 (49%), Gaps = 11/520 (2%) Frame = +1 Query: 403 SCEIEQLKGGSVKDTSKIKEIYEVEIRQLRNLLNDSEKNRAPLEVQIDSLEYELDDLRKR 582 + +++ L+G KDT ++ +YE E+++ R L+ND+ + R LE +I L EL + R++ Sbjct: 26 AADLDLLRGRWGKDTLSVRAMYEGELQEARKLVNDTNRQREELEKEIRKLLDELSEYRRK 85 Query: 583 LMLLEDINGQY--KSTIDKQRQELGNFEGELVELRRFKNTRNADVSKEKEEVKSLRDELK 756 + + G + ID+ +L E E+ LRR ++++ K+E +EL Sbjct: 86 YE--DALRGHQIDRDRIDELLNQLSGLEAEINLLRRRIANIIEEIARIKKENLQFANELT 143 Query: 757 KLRQDYEREALAHITVEHELQNLRETMDFEKQIHETEVKELTTVAYRDPTKENMMYWQSE 936 K R D ++E L I ++++Q L E +DF +++H+ E+ EL +A RD T EN Y+++E Sbjct: 144 KARSDLDQETLNRIDFQNQVQTLLEEIDFMRRVHDQEIAELQAMASRDTTPENREYFKNE 203 Query: 937 MGRALRDIQTEYENKLSGIKQEMEASYGLQMQEIRTSKTRDNVETTHYKEENSRIKSQIQ 1116 + A+RDI+ EY+ + + +ME+ Y L++QEI+T TR N+E + K+ R++ Q+ Sbjct: 204 LASAIRDIRAEYDQICNVNRMDMESWYKLKVQEIQTQSTRQNLEQNYAKK---RLRVQLT 260 Query: 1117 SMRSRIADLEMQNAEYEHNLNMIRGNY--EETQRSLELKLMSXXXXXXXXXXXXSAALTE 1290 +R ++ADLE +N+ + + NY E+ QRS E L A Sbjct: 261 DLRGKLADLEGRNSLLKQTQEL---NYQLEDDQRSYEAALNDRDAQIRKMDEECQA---- 313 Query: 1291 LQSLLDAKLGLELEIAAYAKLLEFEESRVNSNTEGNMIISQFXXXXXXXXXXXXXXXXXX 1470 L LLD K L+ EIA Y K+LE EE+R ++ Sbjct: 314 LMMLLDTKQTLDAEIAIYRKMLEGEENRAGLRQLVEQVVK-------------------- 353 Query: 1471 XXXXXXXXXXXXXXXXXXXXXXXXXMTERETTDYQSKRMKFRRTTKGNISIAETDLTGSF 1650 + + ET S F+R+ KGN+SI ET G + Sbjct: 354 -----------THGLTQVDETESLRVLKGETASRTS----FQRSAKGNVSIQETAPDGRY 398 Query: 1651 IILENI-GTKVESLEGMTIKRAIEGNSNQISYTISSKVTLNAGEKISLVSRGKKTGKGLE 1827 ++LEN +K E++ +KR I+G +I YT+ L G+ + + +RG+ G+ Sbjct: 399 VVLENTHRSKEEAIGEWKLKRKIDG-KREIVYTLPRDFILRPGKSVKIWARGQ---GGIH 454 Query: 1828 IETD------EASWGVGNNVVTTSYDSEGVEKAAHYQEVS 1929 + E ++G G+NV T ++ EG E+A H Q S Sbjct: 455 SPPEQLVFDLEDTFGSGSNVQTILFNREGEERATHIQRSS 494
Score = 42.7 bits (99), Expect = 0.004
Identities = 19/25 (76%), Positives = 22/25 (88%)
Frame = +2
Query: 326 MQDLNERFASYIEKARFLEAQNKQL 400
M DLN+R ASYIEK RFLEAQN++L
Sbjct: 1 MSDLNDRLASYIEKVRFLEAQNRKL 25
>sp|P31732|OV71_ONCVO Muscle cell intermediate filament protein OV71 Length = 432 Score = 200 bits (509), Expect = 1e-50 Identities = 139/460 (30%), Positives = 224/460 (48%), Gaps = 3/460 (0%) Frame = +1 Query: 559 ELDDLRKRLMLLEDINGQYKSTIDKQRQELGNFEGELVELRRFKNTRNADVSKEKEEVKS 738 EL++ +KR + + ID +L N E E+ LRR + +VS+ K++ Sbjct: 5 ELEEYKKRYDEALKAHQADREVIDDLLVKLSNLEAEINLLRRRVSNLEEEVSRIKKDNLY 64 Query: 739 LRDELKKLRQDYEREALAHITVEHELQNLRETMDFEKQIHETEVKELTTVAYRDPTKENM 918 +EL K R + ++E L I ++++Q L E +DF +++H+ E+ EL +A RD T EN Sbjct: 65 FVNELNKARANLDQETLNRIDFQNQVQTLLEEIDFMRRVHDQEISELQAMAARDTTSENR 124 Query: 919 MYWQSEMGRALRDIQTEYENKLSGIKQEMEASYGLQMQEIRTSKTRDNVETTHYKEENSR 1098 Y+++E+ A+RDI+ EY+ + + +ME+ Y L++QEI+T TR N+E + KEE R Sbjct: 125 EYFKNELSSAIRDIRAEYDQICNMNRTDMESWYKLKVQEIQTQSTRQNLEQGYAKEEVKR 184 Query: 1099 IKSQIQSMRSRIADLEMQNAEYEHNLNMIRGNYEETQRSLELKLMSXXXXXXXXXXXXSA 1278 ++ Q+ +R ++ADLE +N+ E + + E+ QRS E L A Sbjct: 185 LRVQLSDLRGKLADLEGRNSLLEKQMQELNYQLEDDQRSYEAALNDRDAQIRKMREECQA 244 Query: 1279 ALTELQSLLDAKLGLELEIAAYAKLLEFEESRVNSNTEGNMIISQFXXXXXXXXXXXXXX 1458 + ELQ LLD K L+ EIA Y K+LE EE+R ++ Sbjct: 245 LMMELQMLLDTKQTLDAEIAIYRKMLEGEENRAGLRQLVEQVVK---------------- 288 Query: 1459 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXMTERETTDYQSKRMKFRRTTKGNISIAETDL 1638 + + ET S F+R+ KGN+SI + Sbjct: 289 ---------------THGLSEIGETESIRVLKGETASRTS----FQRSAKGNVSIQDASS 329 Query: 1639 TGSFIILENI-GTKVESLEGMTIKRAIEGNSNQISYTISSKVTLNAGEKISLVSRGKKTG 1815 G FI+LEN +K E + +KR I+G +I YT L G+ + + +RG+ Sbjct: 330 DGKFILLENTHRSKEEPIGEWRLKRKIDG-KREIVYTFPRDFILKPGKTVKIWARGQGVY 388 Query: 1816 KGLE--IETDEASWGVGNNVVTTSYDSEGVEKAAHYQEVS 1929 + + E S+GVG+NV T ++ EG E+A+H Q S Sbjct: 389 SPPDQLVFDAEDSFGVGSNVQTILFNKEGEERASHIQRSS 428
>sp|Q19289|IFB1_CAEEL Intermediate filament protein ifb-1 (Intermediate filament protein B1) (IF-B1) (Cel IF B1) Length = 589 Score = 190 bits (483), Expect = 1e-47 Identities = 136/517 (26%), Positives = 249/517 (48%), Gaps = 12/517 (2%) Frame = +1 Query: 409 EIEQLKGGSVKDTSKIKEIYEVEIRQLRNLLNDSEKNRAPLEVQIDSLEYELDDLRKRLM 588 ++++L+G KDTS+IK Y + R ++D+ + +A ++V++ +L + R R Sbjct: 112 DLDELRGKWGKDTSEIKIKYSESLSTARKDIDDAARRKAEVDVKVARHRDDLAEYRSRYE 171 Query: 589 LLEDINGQYKSTIDKQRQELGNFEGELVELR-RFKNTRNADVSKEKEEVKSLRDELKKLR 765 ++ + I + + + + E+ LR RFK + + + + + +EL+K R Sbjct: 172 DIQQRRESDREKISQWTNAIADAQSEVEMLRARFKQLTDEE-KRVTADNSRIWEELQKAR 230 Query: 766 QDYEREALAHITVEHELQNLRETMDFEKQIHETEVKELTTVAYRDPTKENMMYWQSEMGR 945 D + E + I ++++Q L E ++F +++HE EVKEL + + P + ++++E+ Sbjct: 231 SDLDDETIGRIDFQNQVQTLMEELEFLRRVHEQEVKELQALLAQAPA-DTREFFKNELAL 289 Query: 946 ALRDIQTEYENKLSGIKQEMEASYGLQMQEIRTSKTRDNVETTHYKEENSRIKSQIQSMR 1125 A+RDI+ EY+ KQ+ME+ Y L++ E++ S R N+E+T+ ++E R++ I +R Sbjct: 290 AIRDIKDEYDIIAKQGKQDMESWYKLKVSEVQGSANRANMESTYQRDEVKRMRDNIGDLR 349 Query: 1126 SRIADLEMQNAEYEHNLNMIRGNYEETQRSLELKLMSXXXXXXXXXXXXSAALTELQSLL 1305 ++ DLE +N+ E + + + QR E L + ELQ+LL Sbjct: 350 GKLGDLENKNSLLEKEVQNLNYQLTDDQRQYEAALNDRDATLRRMREECQTLVAELQALL 409 Query: 1306 DAKLGLELEIAAYAKLLEFEESRVNSNTEGNMIISQFXXXXXXXXXXXXXXXXXXXXXXX 1485 D K L+ EIA Y K+LE EE+RV + Sbjct: 410 DTKQMLDAEIAIYRKMLEGEETRVGLTQMVEQAVK------------------------- 444 Query: 1486 XXXXXXXXXXXXXXXXXXXXMTERETTDYQ-------SKRMKFRRTTKGNISIAETDLTG 1644 + ++E TD S + F+R+ KGN++I+E D G Sbjct: 445 -----------------THSLQQQENTDSTRSVRGEVSTKTTFQRSAKGNVTISECDPNG 487 Query: 1645 SFIILENI-GTKVESLEGMTIKRAIEGNSNQISYTISSKVTLNAGEKISLVSR---GKKT 1812 FI LEN K E++ I+R ++G +I Y+I + V + G+ +++ +R G Sbjct: 488 KFIKLENSHRNKDENVGEHKIRRKLDGR-REIVYSIPANVVIKPGKNLTIYARDQGGINN 546 Query: 1813 GKGLEIETDEASWGVGNNVVTTSYDSEGVEKAAHYQE 1923 + E +WG+G NVVT+ + +G E+A H Q+ Sbjct: 547 PPESLVFDGENTWGIGANVVTSLVNKDGEERATHTQK 583
Score = 45.4 bits (106), Expect = 6e-04
Identities = 28/97 (28%), Positives = 50/97 (51%), Gaps = 9/97 (9%)
Frame = +2
Query: 140 YKPISQPRNNLRDLQKTAGISLNAQMSDQQGGSMVPLIR--------GDSVALQSNV-NS 292
Y+ QPR +R + +G ++ GG ++ ++ G S AL +N S
Sbjct: 14 YRSTIQPRTAVRSQSRQSGNYVSGGNGAGSGGRVLKMVTEMGSATVGGISPALSANAAKS 73
Query: 293 MRKGRAQEKRDMQDLNERFASYIEKARFLEAQNKQLV 403
+ +EK+ +Q LN+R +YI++ + LE QN++LV
Sbjct: 74 FLEATDKEKKTLQGLNDRLGNYIDRVKKLEEQNRKLV 110
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 168,173,584
Number of Sequences: 369166
Number of extensions: 2894338
Number of successful extensions: 13119
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 10455
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 12279
length of database: 68,354,980
effective HSP length: 117
effective length of database: 46,740,985
effective search space used: 25941246675
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail