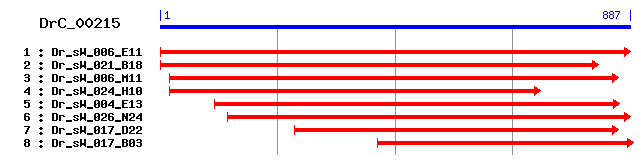
DrC_00215
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00215 (887 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O70352|CD82_RAT CD82 antigen (Metastasis suppressor homo... 60 8e-09 sp|P40237|CD82_MOUSE CD82 antigen (Inducible membrane prote... 60 1e-08 sp|Q80WR1|TSN18_MOUSE Tetraspanin-18 (Tspan-18) 56 2e-07 sp|Q58CY8|TSN18_BOVIN Tetraspanin-18 (Tspan-18) 55 3e-07 sp|P28648|CD63_RAT CD63 antigen (AD1 antigen) 53 1e-06 sp|Q96SJ8|TSN18_HUMAN Tetraspanin-18 (Tspan-18) 52 2e-06 sp|P41731|CD63_MOUSE CD63 antigen 51 5e-06 sp|O60635|TSN1_HUMAN Tetraspanin-1 (Tspan-1) (Tetraspan NET... 51 5e-06 sp|Q5RC27|TSN1_PONPY Tetraspanin-1 (Tspan-1) 49 2e-05 sp|O75954|TSN9_HUMAN Tetraspanin-9 (Tspan-9) (Tetraspan NET-5) 48 4e-05
>sp|O70352|CD82_RAT CD82 antigen (Metastasis suppressor homolog) Length = 266 Score = 60.1 bits (144), Expect = 8e-09 Identities = 56/257 (21%), Positives = 104/257 (40%), Gaps = 2/257 (0%) Frame = +1 Query: 43 GAVILGVGIWVKVDKSNLFEVIRNIKLDDTTASFNNTNSTILDVVAIVLIIXXXXXXXXX 222 GAVILG G+W+ DKS+ V++ T+S+ L V A V I Sbjct: 24 GAVILGFGVWILADKSSFISVLQ-------------TSSSSLQVGAYVFIGVGAITMLMG 70 Query: 223 XXXXXXAIKENKCLLITYSILXXXXXXXXXXXXXXXXXEKNKFQKNLGKSLTNVLEKDFN 402 A+ E +CLL Y + NK ++ +G ++ ++++ N Sbjct: 71 FLGCIGAVNEVRCLLGLYFVFLLLILIAQVTVEVLFYFNANKLKQEMGNTVMDIIQ---N 127 Query: 403 GILNTNATLFTKIFTAIMFELKCCGVNNVTDFDSVRNW--KSTSSSGTKQIYPVGCCKLK 576 +N +++ + + + ++KCCG + S NW + TK YP C K K Sbjct: 128 YSVNASSSR-EEAWDYVQAQVKCCG------WVSPSNWTRNPVLKNSTKTTYPCSCEKTK 180 Query: 577 EKYQYDPNKKFEYNDAKKNFENDNCPFDNDKSTININGCIPSLIDLIEKNMXXXXXXXXX 756 E+ KK + ++ + ++N P D ++ GC+ +++N Sbjct: 181 EEDNQLIVKK-GFCESDNSTASENSPED---WPVHPEGCMEKAQAWLQENFGILLGVCAG 236 Query: 757 XXXFEIMCIVFACCVMR 807 E++ + + C+ R Sbjct: 237 VAVIELLGLFLSICLCR 253
>sp|P40237|CD82_MOUSE CD82 antigen (Inducible membrane protein R2) (C33 antigen) (IA4) Length = 266 Score = 59.7 bits (143), Expect = 1e-08 Identities = 58/258 (22%), Positives = 100/258 (38%), Gaps = 3/258 (1%) Frame = +1 Query: 43 GAVILGVGIWVKVDKSNLFEVIRNIKLDDTTASFNNTNSTILDVVAIVLIIXXXXXXXXX 222 GAVILG G+W+ DK++ V++ T+S+ L V A V I Sbjct: 24 GAVILGFGVWILADKNSFISVLQ-------------TSSSSLQVGAYVFIGVGAITIVMG 70 Query: 223 XXXXXXAIKENKCLLITYSILXXXXXXXXXXXXXXXXXEKNKFQKNLGKSLTNVLEKDFN 402 A+ E +CLL Y + +K +K +G ++ +++ Sbjct: 71 FLGCIGAVNEVRCLLGLYFVFLLLILIAQVTVGVLFYFNADKLKKEMGNTVMDIIRN--- 127 Query: 403 GILNTNATLF-TKIFTAIMFELKCCGVNNVTDFDSVRNWKSTSS--SGTKQIYPVGCCKL 573 NAT + + + ++KCCG + S NW TK YP C K+ Sbjct: 128 --YTANATSSREEAWDYVQAQVKCCG------WVSHYNWTENEELMGFTKTTYPCSCEKI 179 Query: 574 KEKYQYDPNKKFEYNDAKKNFENDNCPFDNDKSTININGCIPSLIDLIEKNMXXXXXXXX 753 KE+ KK + +A + ++N P D +N GC+ +++N Sbjct: 180 KEEDNQLIVKK-GFCEADNSTVSENNPED---WPVNTEGCMEKAQAWLQENFGILLGVCA 235 Query: 754 XXXXFEIMCIVFACCVMR 807 E++ + + C+ R Sbjct: 236 GVAVIELLGLFLSICLCR 253
>sp|Q80WR1|TSN18_MOUSE Tetraspanin-18 (Tspan-18) Length = 248 Score = 55.8 bits (133), Expect = 2e-07 Identities = 53/256 (20%), Positives = 100/256 (39%), Gaps = 1/256 (0%) Frame = +1 Query: 43 GAVILGVGIWVKVDKSNLFEVIRNIKLDDTTASFNNTNSTILDVVAIVLIIXXXXXXXXX 222 GA +LGVGIWV VD + E++ + +L A +++ Sbjct: 24 GACLLGVGIWVLVDPTGFREIV--------------ATNPLLTTGAYIVLAMGGLLFLLG 69 Query: 223 XXXXXXAIKENKCLLITYSILXXXXXXXXXXXXXXXXXEKNKFQKNLGKS-LTNVLEKDF 399 A++EN+CLL+ + + F+++L + T L K + Sbjct: 70 FLGCCGAVRENRCLLLFFFLFILIIFLVELSAAILAFI----FREHLTREFFTKELTKHY 125 Query: 400 NGILNTNATLFTKIFTAIMFELKCCGVNNVTDFDSVRNWKSTSSSGTKQIYPVGCCKLKE 579 G + + +F+ + ++M CCGVN DF ++ + T+++ P CC+ Sbjct: 126 QG--DNDTDVFSATWNSVMITFGCCGVNGPEDFKLASVFRLLTLD-TEEV-PKACCR--- 178 Query: 580 KYQYDPNKKFEYNDAKKNFENDNCPFDNDKSTININGCIPSLIDLIEKNMXXXXXXXXXX 759 +P + +++ + PF IN GC +++ E + Sbjct: 179 ---REPQTRDGVVLSREECQLGRNPF------INKQGCYTVILNTFETYVYLAGAFAIGV 229 Query: 760 XXFEIMCIVFACCVMR 807 E+ +VFA C+ R Sbjct: 230 LAIELFLMVFAMCLFR 245
>sp|Q58CY8|TSN18_BOVIN Tetraspanin-18 (Tspan-18) Length = 249 Score = 54.7 bits (130), Expect = 3e-07 Identities = 54/256 (21%), Positives = 94/256 (36%), Gaps = 1/256 (0%) Frame = +1 Query: 43 GAVILGVGIWVKVDKSNLFEVIRNIKLDDTTASFNNTNSTILDVVAIVLIIXXXXXXXXX 222 GA +LG+GIWV VD + E++ + +L A +L+ Sbjct: 24 GACLLGIGIWVMVDPTGFREIV--------------AANPLLITGAYILLAMGGLLFLLG 69 Query: 223 XXXXXXAIKENKCLLITYSILXXXXXXXXXXXXXXXXXEKNKFQKNLGKS-LTNVLEKDF 399 A++ENKCLL+ + + F+ NL + T L K + Sbjct: 70 FLGCCGAVRENKCLLLFFFLFILIIFLAELSAAILAFI----FRGNLTREFFTKELTKHY 125 Query: 400 NGILNTNATLFTKIFTAIMFELKCCGVNNVTDFDSVRNWKSTSSSGTKQIYPVGCCKLKE 579 G +T+ +F+ + ++M CCGVN DF ++ + + P CC+ +E Sbjct: 126 QGSNDTD--VFSATWNSVMITFGCCGVNGPEDFKYASVFRLLTLDSDE--VPEACCR-RE 180 Query: 580 KYQYDPNKKFEYNDAKKNFENDNCPFDNDKSTININGCIPSLIDLIEKNMXXXXXXXXXX 759 D + C D +N GC +++ E + Sbjct: 181 PQSRD----------GVLLSREECLLGRD-LFLNKQGCYTVILNAFETYVYLAGALAIGV 229 Query: 760 XXFEIMCIVFACCVMR 807 E+ ++FA C+ R Sbjct: 230 LAIELFAMIFAMCLFR 245
>sp|P28648|CD63_RAT CD63 antigen (AD1 antigen) Length = 238 Score = 53.1 bits (126), Expect = 1e-06 Identities = 46/193 (23%), Positives = 74/193 (38%), Gaps = 4/193 (2%) Frame = +1 Query: 241 AIKENKCLLITY----SILXXXXXXXXXXXXXXXXXEKNKFQKNLGKSLTNVLEKDFNGI 408 A KEN CL+IT+ S++ K++F K+ K + N L Sbjct: 75 ACKENYCLMITFAIFLSLIMLVEVAVAIAGYVFRDQVKSEFSKSFQKQMQNYLTD----- 129 Query: 409 LNTNATLFTKIFTAIMFELKCCGVNNVTDFDSVRNWKSTSSSGTKQIYPVGCCKLKEKYQ 588 N AT+ K+ E KCCG +N TD++ + K P CC Sbjct: 130 -NKTATILDKL----QKENKCCGASNYTDWERIPGM-------AKDRVPDSCC------- 170 Query: 589 YDPNKKFEYNDAKKNFENDNCPFDNDKSTININGCIPSLIDLIEKNMXXXXXXXXXXXXF 768 N C D +STI+ GC+ ++ + KN+ Sbjct: 171 -------------INI-TVGCGNDFKESTIHTQGCVETIAAWLRKNVLLVAGAALGIAFV 216 Query: 769 EIMCIVFACCVMR 807 E++ I+F+CC+++ Sbjct: 217 EVLGIIFSCCLVK 229
>sp|Q96SJ8|TSN18_HUMAN Tetraspanin-18 (Tspan-18) Length = 248 Score = 52.4 bits (124), Expect = 2e-06 Identities = 52/256 (20%), Positives = 92/256 (35%), Gaps = 1/256 (0%) Frame = +1 Query: 43 GAVILGVGIWVKVDKSNLFEVIRNIKLDDTTASFNNTNSTILDVVAIVLIIXXXXXXXXX 222 GA +L +GIWV VD + E++ + +L A +L+ Sbjct: 24 GACLLAIGIWVMVDPTGFREIV--------------AANPLLLTGAYILLAMGGLLFLLG 69 Query: 223 XXXXXXAIKENKCLLITYSILXXXXXXXXXXXXXXXXXEKNKFQKNLGKSL-TNVLEKDF 399 A++ENKCLL+ + + F++NL + T L K + Sbjct: 70 FLGCCGAVRENKCLLLFFFLFILIIFLAELSAAILAFI----FRENLTREFFTKELTKHY 125 Query: 400 NGILNTNATLFTKIFTAIMFELKCCGVNNVTDFDSVRNWKSTSSSGTKQIYPVGCCKLKE 579 G N + +F+ + ++M CCGVN DF ++ + + P CC+ +E Sbjct: 126 QG--NNDTDVFSATWNSVMITFGCCGVNGPEDFKFASVFRLLTLDSEE--VPEACCR-RE 180 Query: 580 KYQYDPNKKFEYNDAKKNFENDNCPFDNDKSTININGCIPSLIDLIEKNMXXXXXXXXXX 759 D + C +N GC +++ E + Sbjct: 181 PQSRD----------GVLLSREECLLGR-SLFLNKQGCYTVILNTFETYVYLAGALAIGV 229 Query: 760 XXFEIMCIVFACCVMR 807 E+ ++FA C+ R Sbjct: 230 LAIELFAMIFAMCLFR 245
>sp|P41731|CD63_MOUSE CD63 antigen Length = 238 Score = 50.8 bits (120), Expect = 5e-06 Identities = 44/193 (22%), Positives = 75/193 (38%), Gaps = 4/193 (2%) Frame = +1 Query: 241 AIKENKCLLITY----SILXXXXXXXXXXXXXXXXXEKNKFQKNLGKSLTNVLEKDFNGI 408 A KEN CL+IT+ S++ K++F K+ + + N L+ Sbjct: 75 ACKENYCLMITFAIFLSLIMLVEVAVAIAGYVFRDQVKSEFNKSFQQQMQNYLKD----- 129 Query: 409 LNTNATLFTKIFTAIMFELKCCGVNNVTDFDSVRNWKSTSSSGTKQIYPVGCCKLKEKYQ 588 N AT+ K+ E CCG +N TD++++ K P CC Sbjct: 130 -NKTATILDKL----QKENNCCGASNYTDWENIPGM-------AKDRVPDSCC------- 170 Query: 589 YDPNKKFEYNDAKKNFENDNCPFDNDKSTININGCIPSLIDLIEKNMXXXXXXXXXXXXF 768 N C D +STI+ GC+ ++ + KN+ Sbjct: 171 -------------INI-TVGCGNDFKESTIHTQGCVETIAIWLRKNILLVAAAALGIAFV 216 Query: 769 EIMCIVFACCVMR 807 E++ I+F+CC+++ Sbjct: 217 EVLGIIFSCCLVK 229
>sp|O60635|TSN1_HUMAN Tetraspanin-1 (Tspan-1) (Tetraspan NET-1) (Tetraspanin TM4-C) Length = 241 Score = 50.8 bits (120), Expect = 5e-06 Identities = 49/228 (21%), Positives = 80/228 (35%) Frame = +1 Query: 43 GAVILGVGIWVKVDKSNLFEVIRNIKLDDTTASFNNTNSTILDVVAIVLIIXXXXXXXXX 222 GA +L VGIWV +D ++ ++ L + F N ++ +V + Sbjct: 22 GAALLAVGIWVSIDGASFLKIFG--PLSSSAMQFVNVGYFLIAAGVVVFALGFLGCYG-- 77 Query: 223 XXXXXXAIKENKCLLITYSILXXXXXXXXXXXXXXXXXEKNKFQKNLGKSLTNVLEKDFN 402 A E+KC L+T+ + + L + ++KD+ Sbjct: 78 ------AKTESKCALVTFFFILLLIFIAEVAAAVVALVYTTMAEHFLTLLVVPAIKKDYG 131 Query: 403 GILNTNATLFTKIFTAIMFELKCCGVNNVTDFDSVRNWKSTSSSGTKQIYPVGCCKLKEK 582 + FT+++ M LKCCG N TDF+ +K S+ +P CC Sbjct: 132 SQED-----FTQVWNTTMKGLKCCGFTNYTDFEDSPYFKENSA------FPPFCC----- 175 Query: 583 YQYDPNKKFEYNDAKKNFENDNCPFDNDKSTININGCIPSLIDLIEKN 726 ND N N+ C + GC L+ I N Sbjct: 176 -----------NDNVTNTANETCTKQKAHDQ-KVEGCFNQLLYDIRTN 211
>sp|Q5RC27|TSN1_PONPY Tetraspanin-1 (Tspan-1) Length = 241 Score = 48.9 bits (115), Expect = 2e-05 Identities = 49/228 (21%), Positives = 79/228 (34%) Frame = +1 Query: 43 GAVILGVGIWVKVDKSNLFEVIRNIKLDDTTASFNNTNSTILDVVAIVLIIXXXXXXXXX 222 GA +L VGIWV +D ++ ++ L + F N ++ +V + Sbjct: 22 GAALLAVGIWVSIDGASFLKIFG--PLSSSAMQFVNVGYFLIAAGVVVFALGFLGCYG-- 77 Query: 223 XXXXXXAIKENKCLLITYSILXXXXXXXXXXXXXXXXXEKNKFQKNLGKSLTNVLEKDFN 402 A E+KC L+T+ + + L + ++KD+ Sbjct: 78 ------AKTESKCALMTFFFILLLIFIAEVAAAVVALVYTTMAEHFLTLLVVPAIKKDYG 131 Query: 403 GILNTNATLFTKIFTAIMFELKCCGVNNVTDFDSVRNWKSTSSSGTKQIYPVGCCKLKEK 582 + FT+++ M LKCCG N TDF+ K + +P CC Sbjct: 132 SQED-----FTQVWNTTMKGLKCCGFTNYTDFEDSPYLKENHA------FPPFCC----- 175 Query: 583 YQYDPNKKFEYNDAKKNFENDNCPFDNDKSTININGCIPSLIDLIEKN 726 ND N ND C + + GC L+ I N Sbjct: 176 -----------NDNITNTANDTCTKQKAEDQ-KVEGCFNQLLYDIRTN 211
>sp|O75954|TSN9_HUMAN Tetraspanin-9 (Tspan-9) (Tetraspan NET-5) Length = 239 Score = 47.8 bits (112), Expect = 4e-05 Identities = 52/201 (25%), Positives = 80/201 (39%), Gaps = 14/201 (6%) Frame = +1 Query: 43 GAVILGVGIWVKVDKSNLFEVIRNIKLDDTTASFNNTNSTILDVVAIVLIIXXXXXXXXX 222 G +LGVGIW+ V + N + SF + ++ L V+AI I+ Sbjct: 24 GCGLLGVGIWLSVSQGNFATF---------SPSFPSLSAANL-VIAIGTIVMVTGFLGCL 73 Query: 223 XXXXXXAIKENKCLLITYSILXXXXXXXXXXXXXXXXXEKNKFQKNLGKSLTN--VLEKD 396 AIKENKCLL+++ I+ +K +N K L +L Sbjct: 74 G-----AIKENKCLLLSFFIVLLVILLAELILLILFFVYMDKVNENAKKDLKEGLLLYHT 128 Query: 397 FNGILNTNATLFTKIFTAIMFELKCCGVNNVTDFDSV------------RNWKSTSSSGT 540 N + NA + I E++CCGV + TD+ V N + + T Sbjct: 129 ENNVGLKNA------WNIIQAEMRCCGVTDYTDWYPVLGENTVPDRCCMENSQGCGRNAT 182 Query: 541 KQIYPVGCCKLKEKYQYDPNK 603 ++ GC + K K +D NK Sbjct: 183 TPLWRTGCYE-KVKMWFDDNK 202
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 89,559,936
Number of Sequences: 369166
Number of extensions: 1719341
Number of successful extensions: 4459
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 4262
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4422
length of database: 68,354,980
effective HSP length: 110
effective length of database: 48,034,130
effective search space used: 8886314050
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail