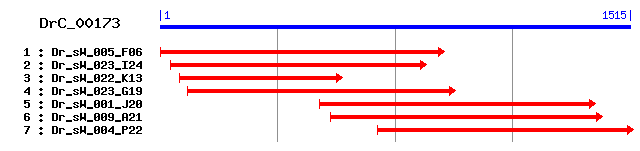
DrC_00173
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00173 (1515 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q7TMR0|PCP_MOUSE Lysosomal Pro-X carboxypeptidase precur... 350 6e-96 sp|Q5RBU7|PCP_PONPY Lysosomal Pro-X carboxypeptidase precur... 342 1e-93 sp|P42785|PCP_HUMAN Lysosomal Pro-X carboxypeptidase precur... 342 2e-93 sp|P34676|YO26_CAEEL Putative serine protease Z688.6 precursor 288 4e-77 sp|Q9EPB1|DPP2_RAT Dipeptidyl-peptidase II precursor (DPP I... 287 5e-77 sp|Q9UHL4|DPP2_HUMAN Dipeptidyl-peptidase II precursor (DPP... 286 8e-77 sp|Q9ET22|DPP2_MOUSE Dipeptidyl-peptidase II precursor (DPP... 284 4e-76 sp|P34610|PCP1_CAEEL Putative serine protease pcp-1 precursor 216 1e-55 sp|Q9NQE7|TSSP_HUMAN Thymus-specific serine protease precursor 103 1e-21 sp|P34528|YM67_CAEEL Putative serine protease K12H4.7 precu... 101 7e-21
>sp|Q7TMR0|PCP_MOUSE Lysosomal Pro-X carboxypeptidase precursor (Prolylcarboxypeptidase) (PRCP) (Proline carboxypeptidase) Length = 491 Score = 350 bits (898), Expect = 6e-96 Identities = 177/428 (41%), Positives = 254/428 (59%), Gaps = 5/428 (1%) Frame = +1 Query: 187 QYETKYFPTYLDHFTYKNDSNKFLMKYLISTKNFVE-GNPILFYCGNEGSIELFANNSGF 363 +Y YF +DHF + D F +YL++ K++ G ILFY GNEG I F NN+GF Sbjct: 45 KYSVLYFEQKVDHFGFA-DMRTFKQRYLVADKHWQRNGGSILFYTGNEGDIVWFCNNTGF 103 Query: 364 VWELGEQLSAIVVFAEHRFYGSTLPFGKKSYDSAQYFGYLNSEQXXXXXXXXXXXXKYNL 543 +W++ E+L A++VFAEHR+YG +LPFG+ S+ +Q+ +L SEQ + + Sbjct: 104 MWDVAEELKAMLVFAEHRYYGESLPFGQDSFKDSQHLNFLTSEQALADFAELIRHLEKTI 163 Query: 544 PGASHSPVIAFGGSYGGMLAAWFRQKYPNIVAGSLAASAPVLMVGNIENCSYPFQVLTKA 723 PGA PVIA GGSYGGMLAAWFR KYP+IV G+LAASAP+ + + C +++T Sbjct: 164 PGAQGQPVIAIGGSYGGMLAAWFRMKYPHIVVGALAASAPIWQLDGMVPCGEFMKIVTND 223 Query: 724 YQTKGSDACVSNVRNVWPVIQQMNDS-LHLVNLSRIFHTCQPL--KNVDDLIAVLTDGLF 894 ++ K C ++R W VI +++ S L +L+ I H C PL + + L + + Sbjct: 224 FR-KSGPYCSESIRKSWNVIDKLSGSGSGLQSLTNILHLCSPLTSEKIPTLKGWIAETWV 282 Query: 895 DMGMANYPYPANFLGPMPAWPVKEFCKPLSNLIKDPIELLNAFFTGLQVFYNYTGQIQCL 1074 ++ M NYPY NFL P+PAWP+KE C+ L N LL F L V+YNY+GQ CL Sbjct: 283 NLAMVNYPYACNFLQPLPAWPIKEVCQYLKNPNVSDTVLLQNIFQALSVYYNYSGQAACL 342 Query: 1075 S-TTTNLPGLDDNGWNVQTCTELPMPMCTDGVKDMFYASSFNKQNNDRSCQKSFGVTPRW 1251 + + T L GW+ Q CTE+ MP CT+G+ DMF ++ + C +GV PR Sbjct: 343 NISQTTTSSLGSMGWSFQACTEMVMPFCTNGIDDMFEPFLWDLEKYSNDCFNQWGVKPRP 402 Query: 1252 GWGQTLFWGKNLNAASNIIFSNGLLDPWAAGGVFNSPNPSIEIVTMKNGAHHVDLRSSHP 1431 W T++ GKN+++ SNIIFSNG LDPW+ GGV ++ + + +GAHH+DLR+ + Sbjct: 403 HWMTTMYGGKNISSHSNIIFSNGELDPWSGGGVTRDITDTLVAINIHDGAHHLDLRAHNA 462 Query: 1432 KDTYDIRL 1455 D + L Sbjct: 463 FDPSSVLL 470
>sp|Q5RBU7|PCP_PONPY Lysosomal Pro-X carboxypeptidase precursor (Prolylcarboxypeptidase) (PRCP) (Proline carboxypeptidase) Length = 496 Score = 342 bits (878), Expect = 1e-93 Identities = 174/427 (40%), Positives = 255/427 (59%), Gaps = 5/427 (1%) Frame = +1 Query: 190 YETKYFPTYLDHFTYKNDSNKFLMKYLISTKNFVE-GNPILFYCGNEGSIELFANNSGFV 366 Y YF +DHF + N F +YL++ K + + G ILFY GNEG I F NN+GF+ Sbjct: 48 YSVLYFQQKVDHFGF-NTVKTFNQRYLVADKYWKKNGGSILFYTGNEGDIIWFCNNTGFM 106 Query: 367 WELGEQLSAIVVFAEHRFYGSTLPFGKKSYDSAQYFGYLNSEQXXXXXXXXXXXXKYNLP 546 W++ E+L A++VFAEHR+YG +LPFG ++ +++ +L SEQ K +P Sbjct: 107 WDVAEELKAMLVFAEHRYYGESLPFGDNTFKDSRHLNFLTSEQALADFAELIKHLKRTIP 166 Query: 547 GASHSPVIAFGGSYGGMLAAWFRQKYPNIVAGSLAASAPVLMVGNIENCSYPFQVLTKAY 726 GA + PVIA GGSYGGMLAAWFR KYP++V G+LAASAP+ ++ C +++T + Sbjct: 167 GAENQPVIAIGGSYGGMLAAWFRMKYPHMVVGALAASAPIWQFEDLVPCGVFMKIVTTDF 226 Query: 727 QTKGSDACVSNVRNVWPVIQQM-NDSLHLVNLSRIFHTCQPL--KNVDDLIAVLTDGLFD 897 + G C ++R W I ++ N L L+ H C PL +++ L +++ + Sbjct: 227 RKSGPH-CSESIRRSWDAINRLSNTGSGLQWLTGALHLCSPLTSQDIQHLKDWISETWVN 285 Query: 898 MGMANYPYPANFLGPMPAWPVKEFCKPLSNLIKDPIELLNAFFTGLQVFYNYTGQIQCLS 1077 + M +YPY +NFL P+PAWP+K C+ L N LL F L V+YNY+GQ++CL+ Sbjct: 286 LAMVDYPYASNFLQPLPAWPIKVVCQYLKNPNVSDSLLLQNIFQALNVYYNYSGQVKCLN 345 Query: 1078 -TTTNLPGLDDNGWNVQTCTELPMPMCTDGVKDMFYASSFNKQNNDRSCQKSFGVTPRWG 1254 + T L GW+ Q CTE+ MP CT+GV DMF S+N + C + +GV PR Sbjct: 346 ISETATSSLGTLGWSYQACTEVVMPFCTNGVDDMFEPHSWNLKELSDDCFQQWGVRPRPS 405 Query: 1255 WGQTLFWGKNLNAASNIIFSNGLLDPWAAGGVFNSPNPSIEIVTMKNGAHHVDLRSSHPK 1434 W T++ GKN+++ +NI+FSNG LDPW+ GGV ++ VT+ GAHH+DLR+ + Sbjct: 406 WITTMYGGKNISSHTNIVFSNGELDPWSGGGVTKDITDTLVAVTISEGAHHLDLRTKNAL 465 Query: 1435 DTYDIRL 1455 D + L Sbjct: 466 DPTSVLL 472
>sp|P42785|PCP_HUMAN Lysosomal Pro-X carboxypeptidase precursor (Prolylcarboxypeptidase) (PRCP) (Proline carboxypeptidase) (Angiotensinase C) (Lysosomal carboxypeptidase C) Length = 496 Score = 342 bits (877), Expect = 2e-93 Identities = 174/427 (40%), Positives = 254/427 (59%), Gaps = 5/427 (1%) Frame = +1 Query: 190 YETKYFPTYLDHFTYKNDSNKFLMKYLISTKNFVE-GNPILFYCGNEGSIELFANNSGFV 366 Y YF +DHF + N F +YL++ K + + G ILFY GNEG I F NN+GF+ Sbjct: 48 YSVLYFQQKVDHFGF-NTVKTFNQRYLVADKYWKKNGGSILFYTGNEGDIIWFCNNTGFM 106 Query: 367 WELGEQLSAIVVFAEHRFYGSTLPFGKKSYDSAQYFGYLNSEQXXXXXXXXXXXXKYNLP 546 W++ E+L A++VFAEHR+YG +LPFG S+ +++ +L SEQ K +P Sbjct: 107 WDVAEELKAMLVFAEHRYYGESLPFGDNSFKDSRHLNFLTSEQALADFAELIKHLKRTIP 166 Query: 547 GASHSPVIAFGGSYGGMLAAWFRQKYPNIVAGSLAASAPVLMVGNIENCSYPFQVLTKAY 726 GA + PVIA GGSYGGMLAAWFR KYP++V G+LAASAP+ ++ C +++T + Sbjct: 167 GAENQPVIAIGGSYGGMLAAWFRMKYPHMVVGALAASAPIWQFEDLVPCGVFMKIVTTDF 226 Query: 727 QTKGSDACVSNVRNVWPVIQQM-NDSLHLVNLSRIFHTCQPL--KNVDDLIAVLTDGLFD 897 + G C ++ W I ++ N L L+ H C PL +++ L +++ + Sbjct: 227 RKSGPH-CSESIHRSWDAINRLSNTGSGLQWLTGALHLCSPLTSQDIQHLKDWISETWVN 285 Query: 898 MGMANYPYPANFLGPMPAWPVKEFCKPLSNLIKDPIELLNAFFTGLQVFYNYTGQIQCLS 1077 + M +YPY +NFL P+PAWP+K C+ L N LL F L V+YNY+GQ++CL+ Sbjct: 286 LAMVDYPYASNFLQPLPAWPIKVVCQYLKNPNVSDSLLLQNIFQALNVYYNYSGQVKCLN 345 Query: 1078 -TTTNLPGLDDNGWNVQTCTELPMPMCTDGVKDMFYASSFNKQNNDRSCQKSFGVTPRWG 1254 + T L GW+ Q CTE+ MP CT+GV DMF S+N + C + +GV PR Sbjct: 346 ISETATSSLGTLGWSYQACTEVVMPFCTNGVDDMFEPHSWNLKELSDDCFQQWGVRPRPS 405 Query: 1255 WGQTLFWGKNLNAASNIIFSNGLLDPWAAGGVFNSPNPSIEIVTMKNGAHHVDLRSSHPK 1434 W T++ GKN+++ +NI+FSNG LDPW+ GGV ++ VT+ GAHH+DLR+ + Sbjct: 406 WITTMYGGKNISSHTNIVFSNGELDPWSGGGVTKDITDTLVAVTISEGAHHLDLRTKNAL 465 Query: 1435 DTYDIRL 1455 D + L Sbjct: 466 DPMSVLL 472
>sp|P34676|YO26_CAEEL Putative serine protease Z688.6 precursor Length = 507 Score = 288 bits (736), Expect = 4e-77 Identities = 173/451 (38%), Positives = 243/451 (53%), Gaps = 27/451 (5%) Frame = +1 Query: 181 EFQYETKYFPTYLDHFTYKNDSNKFLMKYLISTKNFVEGNPILFYCGNEGSIELFANNSG 360 +++YE Y +D F + ND +F ++Y ++ ++ G PILFY GNEGS+E FA N+G Sbjct: 38 KYKYEEGYLKAPIDPFAFTNDL-EFDLRYFLNIDHYETGGPILFYTGNEGSLEAFAENTG 96 Query: 361 FVWELGEQLSAIVVFAEHRFYGSTLPFGKKSYDSAQYFGYLNSEQXXXXXXXXXXXXK-Y 537 F+W+L +L A VVF EHRFYG + PF +SY ++ GYL+S+Q K Sbjct: 97 FMWDLAPELKAAVVFVEHRFYGKSQPFKNESYTDIRHLGYLSSQQALADFALSVQFFKNE 156 Query: 538 NLPGASHSPVIAFGGSYGGMLAAWFRQKYPNIVAGSLAASAPVLMV--GNIENCSYPFQV 711 + GA S VIAFGGSYGGML+AWFR KYP+IV G++AASAPV NI Y F + Sbjct: 157 KIKGAQKSAVIAFGGSYGGMLSAWFRIKYPHIVDGAIAASAPVFWFTDSNIPEDVYDF-I 215 Query: 712 LTKAYQTKGSDACVSNVRNVWPVIQQMNDSLHLVNLSRIFHTCQP---LKNVDD---LIA 873 +T+A+ G + + W + ++ S + + P L+N DD L Sbjct: 216 VTRAFLDAGCNR--KAIEKGWIALDELAKSDSGRQYLNVLYKLDPKSKLENKDDIGFLKQ 273 Query: 874 VLTDGLFDMGMANYPYPANFLGPMPAWPVKEFCKPLSNLIKDPIELLNAFFTGLQVFYNY 1053 + + + M M NYPYP +FL +PAWPVKE CK S K E + + ++YNY Sbjct: 274 YIRESMEAMAMVNYPYPTSFLSSLPAWPVKEACKSASQPGKTQEESAEQLYKIVNLYYNY 333 Query: 1054 TGQ--------IQCLSTTTNLPGLDDNGWNVQTCTELPMPMCTDGVKDMFYASS--FNKQ 1203 TG +C S +L D GW QTCTE+ MP+C G + F+ F + Sbjct: 334 TGDKSTHCANAAKCDSAYGSLG--DPLGWPFQTCTEMVMPLCGSGYPNDFFWKDCPFTSE 391 Query: 1204 NNDRSCQKSFG------VTPRWGWGQTLFWGKNLNAASNIIFSNGLLDPWAAGGVFNSP- 1362 C ++F R G F +L +ASNI+FSNG LDPW+ GG +S Sbjct: 392 KYAEFCMQTFSSIHYNKTLLRPLAGGLAFGATSLPSASNIVFSNGYLDPWSGGGYDHSDK 451 Query: 1363 -NPSIEIVTMKNGAHHVDLRSSHPKDTYDIR 1452 S+ V +K GAHH DLR +HP+DT +++ Sbjct: 452 VQGSVISVILKQGAHHYDLRGAHPQDTEEVK 482
>sp|Q9EPB1|DPP2_RAT Dipeptidyl-peptidase II precursor (DPP II) (Dipeptidyl aminopeptidase II) (Quiescent cell proline dipeptidase) (Dipeptidyl peptidase 7) Length = 500 Score = 287 bits (735), Expect = 5e-77 Identities = 175/433 (40%), Positives = 245/433 (56%), Gaps = 11/433 (2%) Frame = +1 Query: 172 SNLEFQYETKYFPTYLDHFTYKNDSNK-FLMKYLISTKNFVEGN-PILFYCGNEGSIELF 345 S L+ + YF Y+DHF +++ SNK F ++L+S K + G PI FY GNEG I Sbjct: 35 SVLDPDFRENYFEQYMDHFNFESFSNKTFGQRFLVSDKFWKMGEGPIFFYTGNEGDIWSL 94 Query: 346 ANNSGFVWELGEQLSAIVVFAEHRFYGSTLPFGKKSYDSAQYFGYLNSEQXXXXXXXXXX 525 ANNSGF+ EL Q A++VFAEHR+YG +LPFG +S Y L EQ Sbjct: 95 ANNSGFIVELAAQQEALLVFAEHRYYGKSLPFGVQSTQRG-YTQLLTVEQALADFAVLLQ 153 Query: 526 XXKYNLPGASHSPVIAFGGSYGGMLAAWFRQKYPNIVAGSLAASAPVLMVGNIENCSYPF 705 ++NL G +P IAFGGSYGGML+A+ R KYP++VAG+LAASAPV+ V + N F Sbjct: 154 ALRHNL-GVQDAPTIAFGGSYGGMLSAYMRMKYPHLVAGALAASAPVIAVAGLGNPDQFF 212 Query: 706 QVLTKAYQTKGSDACVSNVRNVWPVIQQMNDSLHLVNLSRIFHTCQPL---KNVDDLIAV 876 + +T + + S C VR+ + I+ + +S+ F TCQ L K++ L Sbjct: 213 RDVTADFYGQ-SPKCAQAVRDAFQQIKDLFLQGAYDTISQNFGTCQSLSSPKDLTQLFGF 271 Query: 877 LTDGLFDMGMANYPYPANFLGPMPAWPVKEFCKPL---SNLIKDPIELLNAFF--TGLQV 1041 + + M +YPYP NFLGP+PA PVK C+ L I L + +G++ Sbjct: 272 ARNAFTVLAMMDYPYPTNFLGPLPANPVKVGCERLLSEGQRIMGLRALAGLVYNSSGMEP 331 Query: 1042 FYNYTGQIQ-CLSTTTNLPGLDDNGWNVQTCTELPMPMCTDGVKDMFYASSFNKQNNDRS 1218 ++ Q C T G + W+ Q CTE+ + ++ V DMF F+ + + Sbjct: 332 CFDIYQMYQSCADPTGCGTGSNARAWDYQACTEINLTFDSNNVTDMFPEIPFSDELRQQY 391 Query: 1219 CQKSFGVTPRWGWGQTLFWGKNLNAASNIIFSNGLLDPWAAGGVFNSPNPSIEIVTMKNG 1398 C ++GV PR W QT FWG +L AASNIIFSNG LDPWA GG+ + + SI VT++ G Sbjct: 392 CLDTWGVWPRPDWLQTSFWGGDLKAASNIIFSNGDLDPWAGGGIQRNLSTSIIAVTIQGG 451 Query: 1399 AHHVDLRSSHPKD 1437 AHH+DLR+S+ +D Sbjct: 452 AHHLDLRASNSED 464
>sp|Q9UHL4|DPP2_HUMAN Dipeptidyl-peptidase II precursor (DPP II) (Dipeptidyl aminopeptidase II) (Quiescent cell proline dipeptidase) (Dipeptidyl peptidase 7) Length = 492 Score = 286 bits (733), Expect = 8e-77 Identities = 173/435 (39%), Positives = 241/435 (55%), Gaps = 19/435 (4%) Frame = +1 Query: 190 YETKYFPTYLDHFTYKNDSNK-FLMKYLISTKNFVEGN-PILFYCGNEGSIELFANNSGF 363 ++ ++F LDHF ++ NK F ++L+S + +V G PI FY GNEG + FANNSGF Sbjct: 31 FQERFFQQRLDHFNFERFGNKTFPQRFLVSDRFWVRGEGPIFFYTGNEGDVWAFANNSGF 90 Query: 364 VWELGEQLSAIVVFAEHRFYGSTLPFGKKSYDSAQYFGYLNSEQXXXXXXXXXXXXKYNL 543 V EL + A++VFAEHR+YG +LPFG +S + L EQ + +L Sbjct: 91 VAELAAERGALLVFAEHRYYGKSLPFGAQSTQRG-HTELLTVEQALADFAELLRALRRDL 149 Query: 544 PGASHSPVIAFGGSYGGMLAAWFRQKYPNIVAGSLAASAPVLMVGNIENCSYPFQVLTKA 723 GA +P IAFGGSYGGML+A+ R KYP++VAG+LAASAPVL V + + + F+ +T Sbjct: 150 -GAQDAPAIAFGGSYGGMLSAYLRMKYPHLVAGALAASAPVLAVAGLGDSNQFFRDVTAD 208 Query: 724 YQTKGSDACVSNVRNVWPVIQQMNDSLHLVNLSRIFHTCQPLKNVDDLIAVLT---DGLF 894 ++ + S C VR + I+ + + F TCQPL + DL + + Sbjct: 209 FEGQ-SPKCTQGVREAFRQIKDLFLQGAYDTVRWEFGTCQPLSDEKDLTQLFMFARNAFT 267 Query: 895 DMGMANYPYPANFLGPMPAWPVKEFCKPLSNLIKDPIELLNAFFTGLQVF----YNYTGQ 1062 + M +YPYP +FLGP+PA PVK C L + + TGL+ YN +G Sbjct: 268 VLAMMDYPYPTDFLGPLPANPVKVGCDRLLSEAQR--------ITGLRALAGLVYNASGS 319 Query: 1063 IQCLST----------TTNLPGLDDNGWNVQTCTELPMPMCTDGVKDMFYASSFNKQNND 1212 C T G D W+ Q CTE+ + ++ V DMF F + Sbjct: 320 EHCYDIYRLYHSCADPTGCGTGPDARAWDYQACTEINLTFASNNVTDMFPDLPFTDELRQ 379 Query: 1213 RSCQKSFGVTPRWGWGQTLFWGKNLNAASNIIFSNGLLDPWAAGGVFNSPNPSIEIVTMK 1392 R C ++GV PR W T FWG +L AASNIIFSNG LDPWA GG+ + + S+ VT++ Sbjct: 380 RYCLDTWGVWPRPDWLLTSFWGGDLRAASNIIFSNGNLDPWAGGGIRRNLSASVIAVTIQ 439 Query: 1393 NGAHHVDLRSSHPKD 1437 GAHH+DLR+SHP+D Sbjct: 440 GGAHHLDLRASHPED 454
>sp|Q9ET22|DPP2_MOUSE Dipeptidyl-peptidase II precursor (DPP II) (Dipeptidyl aminopeptidase II) (Quiescent cell proline dipeptidase) (Dipeptidyl peptidase 7) Length = 506 Score = 284 bits (727), Expect = 4e-76 Identities = 173/431 (40%), Positives = 246/431 (57%), Gaps = 11/431 (2%) Frame = +1 Query: 178 LEFQYETKYFPTYLDHFTYKNDSNK-FLMKYLISTKNFVEGN-PILFYCGNEGSIELFAN 351 L+ + YF Y+DHF +++ NK F ++L+S K + G PI FY GNEG I FAN Sbjct: 37 LDPDFHENYFEQYMDHFNFESFGNKTFGQRFLVSDKFWKMGEGPIFFYTGNEGDIWSFAN 96 Query: 352 NSGFVWELGEQLSAIVVFAEHRFYGSTLPFGKKSYDSAQYFGYLNSEQXXXXXXXXXXXX 531 NSGF+ EL Q A++VFAEHR+YG +LPFG +S Y L EQ Sbjct: 97 NSGFMVELAAQQEALLVFAEHRYYGKSLPFGVQSTQRG-YTQLLTVEQALADFAVLLQAL 155 Query: 532 KYNLPGASHSPVIAFGGSYGGMLAAWFRQKYPNIVAGSLAASAPVLMVGNIENCSYPFQV 711 + +L G +P IAFGGSYGGML+A+ R KYP++VAG+LAASAPV+ V + + F+ Sbjct: 156 RQDL-GVHDAPTIAFGGSYGGMLSAYMRMKYPHLVAGALAASAPVVAVAGLGDSYQFFRD 214 Query: 712 LTKAYQTKGSDACVSNVRNVWPVIQQMNDSLHLVNLSRIFHTCQPL---KNVDDLIAVLT 882 +T + + S C VR+ + I+ + +S+ F TCQ L K++ L Sbjct: 215 VTADFYGQ-SPKCAQAVRDAFQQIKDLFLQGAYDTISQNFGTCQSLSSPKDLTQLFGFAR 273 Query: 883 DGLFDMGMANYPYPANFLGPMPAWPVKEFCKPLSNLIKDPIEL-----LNAFFTGLQVFY 1047 + + M +YPYP +FLGP+PA PVK C+ L N + + L L +G + Y Sbjct: 274 NAFTVLAMMDYPYPTDFLGPLPANPVKVGCQRLLNEGQRIMGLRALAGLVYNSSGTEPCY 333 Query: 1048 N-YTGQIQCLSTTTNLPGLDDNGWNVQTCTELPMPMCTDGVKDMFYASSFNKQNNDRSCQ 1224 + Y C T G D W+ Q CTE+ + ++ V DMF F+++ + C Sbjct: 334 DIYRLYQSCADPTGCGTGSDARAWDYQACTEINLTFDSNNVTDMFPEIPFSEELRQQYCL 393 Query: 1225 KSFGVTPRWGWGQTLFWGKNLNAASNIIFSNGLLDPWAAGGVFNSPNPSIEIVTMKNGAH 1404 ++GV PR W QT FWG +L AASNIIFSNG LDPWA GG+ ++ + S+ VT++ GAH Sbjct: 394 DTWGVWPRQDWLQTSFWGGDLKAASNIIFSNGDLDPWAGGGIQSNLSTSVIAVTIQGGAH 453 Query: 1405 HVDLRSSHPKD 1437 H+DLR+S+ +D Sbjct: 454 HLDLRASNSED 464
>sp|P34610|PCP1_CAEEL Putative serine protease pcp-1 precursor Length = 565 Score = 216 bits (551), Expect = 1e-55 Identities = 147/444 (33%), Positives = 221/444 (49%), Gaps = 37/444 (8%) Frame = +1 Query: 217 LDHFTYKNDSNKFLMKYLISTKNFVEGNPILFYCGNEGSIELFANNSGFVWELGEQLSAI 396 LDHFT+ D+ F M+ + + + G PI FY GNEG +E F +G +++L +A Sbjct: 51 LDHFTW-GDTRTFDMRVMWNNTFYKPGGPIFFYTGNEGGLESFVTATGMMFDLAPMFNAS 109 Query: 397 VVFAEHRFYGSTLPFGKKSYDSAQYFGYLNSEQXXXXXXXXXXXXK-----YNLPGASHS 561 ++FAEHRFYG T PFG +SY S GYL SEQ K + + + + Sbjct: 110 IIFAEHRFYGQTQPFGNQSYASLANVGYLTSEQALADYAELLTELKRDNNQFKMTFPAAT 169 Query: 562 PVIAFGGSYGGMLAAWFRQKYPNIVAGSLAASAPVLMV--GNIENCSYPFQVLTKAYQTK 735 VI+FGGSYGGML+AWFRQKYP+IV G+ A SAP++ + G ++ ++ + ++ Y Sbjct: 170 QVISFGGSYGGMLSAWFRQKYPHIVKGAWAGSAPLIYMNGGGVDPGAFD-HITSRTYIDN 228 Query: 736 GSDACV-SNVRNVWPVIQQMNDSLHLVNLSRIF--HTCQPLKNVDD---LIAVLTDGLFD 897 G + + +N N + + +N + +F ++N D L A L + + Sbjct: 229 GCNRFILANAWNATLNLSSTDAGRQWLNNNTVFKLDPRTKIRNQTDGWNLNAYLREAIEY 288 Query: 898 MGMANYPYPANFLGPMPAWPVKEFCKPL--SNLIKDPIELLNAFFTGLQVFYNYTGQIQ- 1068 M M +YPYP FL P+PAWPV C + + +L+ A ++YNY Sbjct: 289 MAMVDYPYPTGFLEPLPAWPVTVACGYMNANGTSFSDKDLVKAVANAANIYYNYNRDPNF 348 Query: 1069 --------CLSTTTNLPGLDDNGWNVQTCTELPMPMCTDGVKDMFYASSFNK------QN 1206 C T G D+ GW Q C+E+ M MC G + + + K Q Sbjct: 349 TYCIDFSICGDQGTGGLGGDELGWPWQECSEIIMAMCASGGSNDVFWNECGKDIYQTLQQ 408 Query: 1207 NDRSCQKSFGVTPRWGWG----QTLFWGKNLNAASNIIFSNGLLDPWAAGGV---FNSPN 1365 S KS G TP+ W +TL+ G +L+ +SN+I + G LDPW+ GG N+ Sbjct: 409 GCVSIFKSMGWTPK-NWNIDAVKTLY-GYDLSGSSNLILTQGHLDPWSGGGYKVDQNNAA 466 Query: 1366 PSIEIVTMKNGAHHVDLRSSHPKD 1437 I ++ + AHH+DLR + D Sbjct: 467 RGIYVLEIPGSAHHLDLRQPNTCD 490
>sp|Q9NQE7|TSSP_HUMAN Thymus-specific serine protease precursor Length = 514 Score = 103 bits (258), Expect = 1e-21 Identities = 109/433 (25%), Positives = 175/433 (40%), Gaps = 20/433 (4%) Frame = +1 Query: 217 LDHFTYKNDSNKFLMKYLISTKNFV-EGNPILFYCGNEGSIELFANNSGFVWELGEQLSA 393 LD F +D FL +Y ++ +++V + PI + G EGS+ + G L A Sbjct: 66 LDPFNV-SDRRSFLQRYWVNDQHWVGQDGPIFLHLGGEGSLGPGSVMRGHPAALAPAWGA 124 Query: 394 IVVFAEHRFYGSTLPFGKKSYDSAQYFGYLNSEQXXXXXXXXXXXXKYNLPGASHSPVIA 573 +V+ EHRFYG ++P G + AQ +L+S +S SP I Sbjct: 125 LVISLEHRFYGLSIPAG--GLEMAQ-LRFLSSRLALADVVSARLALSRLFNISSSSPWIC 181 Query: 574 FGGSYGGMLAAWFRQKYPNIVAGSLAASAPVLMVGNIENCSYPFQVLTKAYQTKGSDACV 753 FGGSY G LAAW R K+P+++ S+A+SAPV V + + + GS C Sbjct: 182 FGGSYAGSLAAWARLKFPHLIFASVASSAPVRAVLDFSEYNDVVSRSLMSTAIGGSLECR 241 Query: 754 SNVRNVWPVIQQ--MNDSLHLVNLSRIFHTCQPLKNVDDLIAVLTDGLFDMGMANYPYPA 927 + V + +++ + L C PL ++ A L L + Y Sbjct: 242 AAVSVAFAEVERRLRSGGAAQAALRTELSACGPLGRAENQ-AELLGALQALVGGVVQYDG 300 Query: 928 NFLGPMPAWPVKEFC-------------KPLSNLIKDPIELLNAFFTGLQVFYNYTGQIQ 1068 P+ V++ C P L + +L++ F Q Sbjct: 301 QTGAPL---SVRQLCGLLLGGGGNRSHSTPYCGLRRAVQIVLHSLGQKCLSFSRAETVAQ 357 Query: 1069 CLSTTTNLPGLDDNGWNVQTCTELPMPMCTDGVKDMFYASSFNKQNNDRSCQKSFGVTP- 1245 ST L G+ D W QTCTE + + + F D C++ FG++ Sbjct: 358 LRSTEPQLSGVGDRQWLYQTCTEFGFYVTCENPRCPFSQLPALPSQLD-LCEQVFGLSAL 416 Query: 1246 --RWGWGQT-LFWGKNLNAASNIIFSNGLLDPWAAGGVFNSPNPSIEIVTMKNGAHHVDL 1416 QT ++G A+ ++F NG DPW V + S + ++ G+H +D+ Sbjct: 417 SVAQAVAQTNSYYGGQTPGANKVLFVNGDTDPWHVLSVTQALGSSESTLLIRTGSHCLDM 476 Query: 1417 RSSHPKDTYDIRL 1455 P D+ +RL Sbjct: 477 APERPSDSPSLRL 489
>sp|P34528|YM67_CAEEL Putative serine protease K12H4.7 precursor Length = 510 Score = 101 bits (251), Expect = 7e-21 Identities = 108/444 (24%), Positives = 170/444 (38%), Gaps = 32/444 (7%) Frame = +1 Query: 205 FPTYLDHFTYKNDSNKFLMKYLISTKNFVEGNPILFYCGNEG-SIELFANNSGF-VWELG 378 F LDHF + F +Y + + + G P G EG + + G + L Sbjct: 63 FTQTLDHFD-SSVGKTFQQRYYHNNQWYKAGGPAFLMLGGEGPESSYWVSYPGLEITNLA 121 Query: 379 EQLSAIVVFAEHRFYGSTLPFGKKSYDSAQYFGYLNSEQXXXXXXXXXXXXKYNLPGASH 558 + A V EHRFYG T P S + +Y L+S Q P ++ Sbjct: 122 AKQGAWVFDIEHRFYGETHPTSDMSVPNLKY---LSSAQAIEDAAAFIKAMTAKFPQLAN 178 Query: 559 SPVIAFGGSYGGMLAAWFRQKYPNIVAGSLAASAPVLMVGNIENCSYPFQVLTKAYQ--- 729 + + FGGSY G LAAW R K+P +V ++ +S PV F+ + Q Sbjct: 179 AKWVTFGGSYSGALAAWTRAKHPELVYAAVGSSGPV-------QAEVDFKEYLEVVQNSI 231 Query: 730 TKGSDACVSNVRNVWPVIQQ-MNDSLHLVNLSRIFHTCQPLKNVDDLIAVLTDGLFDMGM 906 T+ S C ++V + ++ + S L FH CQ ++ D + + ++ M Sbjct: 232 TRNSTECAASVTQGFNLVASLLQTSDGRKQLKTAFHLCQDIQMDDKSLKYFWETVYSPYM 291 Query: 907 ANYPYPANFLGPMPAWPV--KEFCKPLSNLIKDPIELL---NAFFTGLQVFY-----NYT 1056 Y + G C+ N P++ L N +F + ++ +Y Sbjct: 292 EVVQYSGDAAGSFATQLTISHAICRYHINTKSTPLQKLKQVNDYFNQVSGYFGCNDIDYN 351 Query: 1057 GQIQCLSTTTNLPGLDDNGWNVQTCTELPMPMCTD---------GVKDM---FY----AS 1188 G I + T D W QTCTE T GV ++ +Y + Sbjct: 352 GFISFMKDETFGEAQSDRAWVWQTCTEFGYYQSTSSATAGPWFGGVSNLPAQYYIDECTA 411 Query: 1189 SFNKQNNDRSCQKSFGVTPRWGWGQTLFWGKNLNAASNIIFSNGLLDPWAAGGVFNSPNP 1368 + N + Q S T ++ G+ NLN I+ NG +DPW A G S N Sbjct: 412 IYGAAYNSQEVQTSVDYTNQYYGGR-----DNLN-TDRILLPNGDIDPWHALGKLTSSNS 465 Query: 1369 SIEIVTMKNGAHHVDLRSSHPKDT 1440 +I V + AH D+ + D+ Sbjct: 466 NIVPVVINGTAHCADMYGASSLDS 489
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 169,520,210
Number of Sequences: 369166
Number of extensions: 3635806
Number of successful extensions: 9556
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9024
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9517
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 18325966995
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail