DrC_00147
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
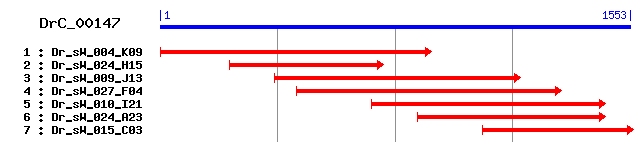
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00147 (1553 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9SZ67|WRK19_ARATH Probable WRKY transcription factor 19... 97 1e-19 sp|Q9FZ36|M3K2_ARATH Mitogen-activated protein kinase kinas... 94 1e-18 sp|Q13043|STK4_HUMAN Serine/threonine-protein kinase 4 (STE... 93 2e-18 sp|O22040|M3K1_ARATH Mitogen-activated protein kinase kinas... 92 5e-18 sp|Q9JI10|STK3_MOUSE Serine/threonine-protein kinase 3 (STE... 91 7e-18 sp|Q13188|STK3_HUMAN Serine/threonine-protein kinase 3 (STE... 91 9e-18 sp|O22042|M3K3_ARATH Mitogen-activated protein kinase kinas... 89 3e-17 sp|Q60700|M3K12_MOUSE Mitogen-activated protein kinase kina... 89 5e-17 sp|Q12852|M3K12_HUMAN Mitogen-activated protein kinase kina... 89 5e-17 sp|P50528|PLO1_SCHPO Serine/threonine-protein kinase plo1 88 6e-17
>sp|Q9SZ67|WRK19_ARATH Probable WRKY transcription factor 19 (WRKY DNA-binding protein 19) Length = 1895 Score = 97.4 bits (241), Expect = 1e-19 Identities = 66/194 (34%), Positives = 101/194 (52%), Gaps = 5/194 (2%) Frame = +2 Query: 911 EEENSVFIVMERMAS-SLRSLLKPDTQHLTEDKIKKYSYEILDGLSYMHKLKILHRSLNC 1087 ++E++++I +E + SLR L + + L + + Y+ +ILDGL Y+H +HR++ C Sbjct: 1695 KDESNLYIFLELVTQGSLRKLYQRN--QLGDSVVSLYTRQILDGLKYLHDKGFIHRNIKC 1752 Query: 1088 GSVLIDCNDNAKLCNFHLSKFLFDCSTEQLLKGVKGSKIEDDECWVAPEVLLQ----EPY 1255 +VL+D N KL +F L+K + T W+APEV+L + Y Sbjct: 1753 ANVLVDANGTVKLADFGLAKVMSLWRTPYW-------------NWMAPEVILNPKDYDGY 1799 Query: 1256 GFKADIWSFGVTMLEMMTGRRPYPQLNNVQAAHRIVADRHIEFDRDLYPNSGFDEILDTC 1435 G ADIWS G T+LEM+TG+ PY L A + I + + D+ D IL TC Sbjct: 1800 GTPADIWSLGCTVLEMLTGQIPYSDLEIGTALYNIGTGKLPKIP-DILSLDARDFIL-TC 1857 Query: 1436 LRSNQNERPNVFDL 1477 L+ N ERP +L Sbjct: 1858 LKVNPEERPTAAEL 1871
>sp|Q9FZ36|M3K2_ARATH Mitogen-activated protein kinase kinase kinase 2 (Arabidospsis NPK1-related protein kinase 2) Length = 651 Score = 94.0 bits (232), Expect = 1e-18 Identities = 61/198 (30%), Positives = 100/198 (50%), Gaps = 2/198 (1%) Frame = +2 Query: 890 VQLLSMYEEENSVFIVMERMASSLRSLLKPDTQHLTEDKIKKYSYEILDGLSYMHKLKIL 1069 V+ L E+ ++ I++E + S L E ++ Y+ ++L GL Y+H I+ Sbjct: 134 VRYLGTVREDETLNILLEFVPGGSISSLLEKFGAFPESVVRTYTNQLLLGLEYLHNHAIM 193 Query: 1070 HRSLNCGSVLIDCNDNAKLCNFHLSKFLFDCSTEQLLKGVKGSKIEDDECWVAPEVLLQE 1249 HR + ++L+D KL +F SK + + +T K +KG+ W+APEV+LQ Sbjct: 194 HRDIKGANILVDNQGCIKLADFGASKQVAELATISGAKSMKGTPY-----WMAPEVILQT 248 Query: 1250 PYGFKADIWSFGVTMLEMMTGRRPYPQLNNVQAA--HRIVADRHIEFDRDLYPNSGFDEI 1423 + F ADIWS G T++EM+TG+ P+ Q AA H H ++ +S ++ Sbjct: 249 GHSFSADIWSVGCTVIEMVTGKAPWSQQYKEIAAIFHIGTTKSHPPIPDNI--SSDANDF 306 Query: 1424 LDTCLRSNQNERPNVFDL 1477 L CL+ N RP +L Sbjct: 307 LLKCLQQEPNLRPTASEL 324
>sp|Q13043|STK4_HUMAN Serine/threonine-protein kinase 4 (STE20-like kinase MST1) (MST-1) (Mammalian STE20-like protein kinase 1) (Serine/threonine-protein kinase Krs-2) Length = 487 Score = 92.8 bits (229), Expect = 2e-18 Identities = 56/202 (27%), Positives = 99/202 (49%), Gaps = 1/202 (0%) Frame = +2 Query: 875 DTSRFVQLLSMYEEENSVFIVMERM-ASSLRSLLKPDTQHLTEDKIKKYSYEILDGLSYM 1051 D+ V+ Y + ++IVME A S+ +++ + LTED+I L GL Y+ Sbjct: 81 DSPHVVKYYGSYFKNTDLWIVMEYCGAGSVSDIIRLRNKTLTEDEIATILQSTLKGLEYL 140 Query: 1052 HKLKILHRSLNCGSVLIDCNDNAKLCNFHLSKFLFDCSTEQLLKGVKGSKIEDDECWVAP 1231 H ++ +HR + G++L++ +AKL +F ++ L D K + + W+AP Sbjct: 141 HFMRKIHRDIKAGNILLNTEGHAKLADFGVAGQLTDTMA-------KRNTVIGTPFWMAP 193 Query: 1232 EVLLQEPYGFKADIWSFGVTMLEMMTGRRPYPQLNNVQAAHRIVADRHIEFDRDLYPNSG 1411 EV+ + Y ADIWS G+T +EM G+ PY ++ ++A I + F + + Sbjct: 194 EVIQEIGYNCVADIWSLGITAIEMAEGKPPYADIHPMRAIFMIPTNPPPTFRKPELWSDN 253 Query: 1412 FDEILDTCLRSNQNERPNVFDL 1477 F + + CL + +R L Sbjct: 254 FTDFVKQCLVKSPEQRATATQL 275
>sp|O22040|M3K1_ARATH Mitogen-activated protein kinase kinase kinase 1 (Arabidospsis NPK1-related protein kinase 1) Length = 666 Score = 91.7 bits (226), Expect = 5e-18 Identities = 59/196 (30%), Positives = 97/196 (49%) Frame = +2 Query: 890 VQLLSMYEEENSVFIVMERMASSLRSLLKPDTQHLTEDKIKKYSYEILDGLSYMHKLKIL 1069 V+ L E++++ I++E + S L E ++ Y+ ++L GL Y+H I+ Sbjct: 135 VRYLGTVREDDTLNILLEFVPGGSISSLLEKFGPFPESVVRTYTRQLLLGLEYLHNHAIM 194 Query: 1070 HRSLNCGSVLIDCNDNAKLCNFHLSKFLFDCSTEQLLKGVKGSKIEDDECWVAPEVLLQE 1249 HR + ++L+D KL +F SK + + +T K +KG+ W+APEV+LQ Sbjct: 195 HRDIKGANILVDNKGCIKLADFGASKQVAELATMTGAKSMKGTPY-----WMAPEVILQT 249 Query: 1250 PYGFKADIWSFGVTMLEMMTGRRPYPQLNNVQAAHRIVADRHIEFDRDLYPNSGFDEILD 1429 + F ADIWS G T++EM+TG+ P+ Q AA + +S + L Sbjct: 250 GHSFSADIWSVGCTVIEMVTGKAPWSQQYKEVAAIFFIGTTKSHPPIPDTLSSDAKDFLL 309 Query: 1430 TCLRSNQNERPNVFDL 1477 CL+ N RP +L Sbjct: 310 KCLQEVPNLRPTASEL 325
>sp|Q9JI10|STK3_MOUSE Serine/threonine-protein kinase 3 (STE20-like kinase MST2) (MST-2) (Mammalian STE20-like protein kinase 2) Length = 497 Score = 91.3 bits (225), Expect = 7e-18 Identities = 56/202 (27%), Positives = 99/202 (49%), Gaps = 1/202 (0%) Frame = +2 Query: 875 DTSRFVQLLSMYEEENSVFIVMERM-ASSLRSLLKPDTQHLTEDKIKKYSYEILDGLSYM 1051 D+ V+ Y + ++IVME A S+ +++ + LTED+I L GL Y+ Sbjct: 78 DSPYVVKYYGSYFKNTDLWIVMEYCGAGSVSDIIRLRNKTLTEDEIATILKSTLKGLEYL 137 Query: 1052 HKLKILHRSLNCGSVLIDCNDNAKLCNFHLSKFLFDCSTEQLLKGVKGSKIEDDECWVAP 1231 H ++ +HR + G++L++ +AKL +F ++ L D K + + W+AP Sbjct: 138 HFMRKIHRDIKAGNILLNTEGHAKLADFGVAGQLTDTMA-------KRNTVIGTPFWMAP 190 Query: 1232 EVLLQEPYGFKADIWSFGVTMLEMMTGRRPYPQLNNVQAAHRIVADRHIEFDRDLYPNSG 1411 EV+ + Y ADIWS G+T +EM G+ PY ++ ++A I + F + + Sbjct: 191 EVIQEIGYNCVADIWSLGITSIEMAEGKPPYADIHPMRAIFMIPTNPPPTFRKPELWSDD 250 Query: 1412 FDEILDTCLRSNQNERPNVFDL 1477 F + + CL + +R L Sbjct: 251 FTDFVKKCLVKSPEQRATATQL 272
>sp|Q13188|STK3_HUMAN Serine/threonine-protein kinase 3 (STE20-like kinase MST2) (MST-2) (Mammalian STE20-like protein kinase 2) (Serine/threonine-protein kinase Krs-1) Length = 491 Score = 90.9 bits (224), Expect = 9e-18 Identities = 56/202 (27%), Positives = 98/202 (48%), Gaps = 1/202 (0%) Frame = +2 Query: 875 DTSRFVQLLSMYEEENSVFIVMERM-ASSLRSLLKPDTQHLTEDKIKKYSYEILDGLSYM 1051 D+ V+ Y + ++IVME A S+ +++ + L ED+I L GL Y+ Sbjct: 78 DSPYVVKYYGSYFKNTDLWIVMEYCGAGSVSDIIRLRNKTLIEDEIATILKSTLKGLEYL 137 Query: 1052 HKLKILHRSLNCGSVLIDCNDNAKLCNFHLSKFLFDCSTEQLLKGVKGSKIEDDECWVAP 1231 H ++ +HR + G++L++ +AKL +F ++ L D K + + W+AP Sbjct: 138 HFMRKIHRDIKAGNILLNTEGHAKLADFGVAGQLTDTMA-------KRNTVIGTPFWMAP 190 Query: 1232 EVLLQEPYGFKADIWSFGVTMLEMMTGRRPYPQLNNVQAAHRIVADRHIEFDRDLYPNSG 1411 EV+ + Y ADIWS G+T +EM G+ PY ++ ++A I + F + + Sbjct: 191 EVIQEIGYNCVADIWSLGITSIEMAEGKPPYADIHPMRAIFMIPTNPPPTFRKPELWSDD 250 Query: 1412 FDEILDTCLRSNQNERPNVFDL 1477 F + + CL N +R L Sbjct: 251 FTDFVKKCLVKNPEQRATATQL 272
>sp|O22042|M3K3_ARATH Mitogen-activated protein kinase kinase kinase 3 (Arabidospsis NPK1-related protein kinase 3) Length = 651 Score = 89.0 bits (219), Expect = 3e-17 Identities = 62/198 (31%), Positives = 96/198 (48%), Gaps = 2/198 (1%) Frame = +2 Query: 890 VQLLSMYEEENSVFIVMERMASSLRSLLKPDTQHLTEDKIKKYSYEILDGLSYMHKLKIL 1069 V+ L E +S+ I+ME + S L E I Y+ ++L GL Y+H I+ Sbjct: 134 VRYLGTVRESDSLNILMEFVPGGSISSLLEKFGSFPEPVIIMYTKQLLLGLEYLHNNGIM 193 Query: 1070 HRSLNCGSVLIDCNDNAKLCNFHLSKFLFDCSTEQLLKGVKGSKIEDDECWVAPEVLLQE 1249 HR + ++L+D +L +F SK + + +T K +KG+ W+APEV+LQ Sbjct: 194 HRDIKGANILVDNKGCIRLADFGASKKVVELATVNGAKSMKGTPY-----WMAPEVILQT 248 Query: 1250 PYGFKADIWSFGVTMLEMMTGRRPYPQLNNVQAA--HRIVADRHIEFDRDLYPNSGFDEI 1423 + F ADIWS G T++EM TG+ P+ + AA H H DL P + + Sbjct: 249 GHSFSADIWSVGCTVIEMATGKPPWSEQYQQFAAVLHIGRTKAHPPIPEDLSPEA--KDF 306 Query: 1424 LDTCLRSNQNERPNVFDL 1477 L CL + R + +L Sbjct: 307 LMKCLHKEPSLRLSATEL 324
>sp|Q60700|M3K12_MOUSE Mitogen-activated protein kinase kinase kinase 12 (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK) Length = 888 Score = 88.6 bits (218), Expect = 5e-17 Identities = 66/233 (28%), Positives = 105/233 (45%) Frame = +2 Query: 800 DISRTNVKNFKMLKYEMVTRFKLPYDTSRFVQLLSMYEEENSVFIVMERMASSLRSLLKP 979 D+ T++K+ + LK+ + FK + +L + + ++ V+ +P Sbjct: 189 DLKETDIKHLRKLKHPNIITFKGVCTQAPCYCILMEFCAQGQLYEVLRAG--------RP 240 Query: 980 DTQHLTEDKIKKYSYEILDGLSYMHKLKILHRSLNCGSVLIDCNDNAKLCNFHLSKFLFD 1159 T L D +S I G++Y+H KI+HR L ++LI +D K+ +F SK L D Sbjct: 241 VTPSLLVD----WSMGIAGGMNYLHLHKIIHRDLKSPNMLITYDDVVKISDFGTSKELSD 296 Query: 1160 CSTEQLLKGVKGSKIEDDECWVAPEVLLQEPYGFKADIWSFGVTMLEMMTGRRPYPQLNN 1339 ST+ G W+APEV+ EP K DIWSFGV + E++TG PY +++ Sbjct: 297 KSTKMSFAGTVA--------WMAPEVIRNEPVSEKVDIWSFGVVLWELLTGEIPYKDVDS 348 Query: 1340 VQAAHRIVADRHIEFDRDLYPNSGFDEILDTCLRSNQNERPNVFDLKLLFHVA 1498 A V + GF +L C S RP+ + L +A Sbjct: 349 -SAIIWGVGSNSLHLPVPSSCPDGFKILLRQCWNSKPRNRPSFRQILLHLDIA 400
>sp|Q12852|M3K12_HUMAN Mitogen-activated protein kinase kinase kinase 12 (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK) Length = 859 Score = 88.6 bits (218), Expect = 5e-17 Identities = 66/233 (28%), Positives = 105/233 (45%) Frame = +2 Query: 800 DISRTNVKNFKMLKYEMVTRFKLPYDTSRFVQLLSMYEEENSVFIVMERMASSLRSLLKP 979 D+ T++K+ + LK+ + FK + +L + + ++ V+ +P Sbjct: 156 DLKETDIKHLRKLKHPNIITFKGVCTQAPCYCILMEFCAQGQLYEVLRAG--------RP 207 Query: 980 DTQHLTEDKIKKYSYEILDGLSYMHKLKILHRSLNCGSVLIDCNDNAKLCNFHLSKFLFD 1159 T L D +S I G++Y+H KI+HR L ++LI +D K+ +F SK L D Sbjct: 208 VTPSLLVD----WSMGIAGGMNYLHLHKIIHRDLKSPNMLITYDDVVKISDFGTSKELSD 263 Query: 1160 CSTEQLLKGVKGSKIEDDECWVAPEVLLQEPYGFKADIWSFGVTMLEMMTGRRPYPQLNN 1339 ST+ G W+APEV+ EP K DIWSFGV + E++TG PY +++ Sbjct: 264 KSTKMSFAGTVA--------WMAPEVIRNEPVSEKVDIWSFGVVLWELLTGEIPYKDVDS 315 Query: 1340 VQAAHRIVADRHIEFDRDLYPNSGFDEILDTCLRSNQNERPNVFDLKLLFHVA 1498 A V + GF +L C S RP+ + L +A Sbjct: 316 -SAIIWGVGSNSLHLPVPSSCPDGFKILLRQCWNSKPRNRPSFRQILLHLDIA 367
>sp|P50528|PLO1_SCHPO Serine/threonine-protein kinase plo1 Length = 683 Score = 88.2 bits (217), Expect = 6e-17 Identities = 58/230 (25%), Positives = 121/230 (52%), Gaps = 5/230 (2%) Frame = +2 Query: 803 ISRTNVKNFKMLKYEMVTRFKLPYDTSR--FVQLLSMYEEENSVFIVMERMA-SSLRSLL 973 I++ +++N K K ++ K+ S V + +E+ ++++++E SL LL Sbjct: 71 IAKRSLQNDKT-KLKLFGEIKVHQSMSHPNIVGFIDCFEDSTNIYLILELCEHKSLMELL 129 Query: 974 KPDTQHLTEDKIKKYSYEILDGLSYMHKLKILHRSLNCGSVLIDCNDNAKLCNFHLSKFL 1153 + Q LTE +++ +IL L YMHK +++HR L G++++D ++N K+ +F L+ L Sbjct: 130 RKRKQ-LTEPEVRYLMMQILGALKYMHKKRVIHRDLKLGNIMLDESNNVKIGDFGLAALL 188 Query: 1154 FDCSTEQLLKGVKGSKIEDDECWVAPEVLL--QEPYGFKADIWSFGVTMLEMMTGRRPYP 1327 D ++ I ++APE+L +E + F+ D+WS GV M ++ G+ P+ Sbjct: 189 MDDEERKM-------TICGTPNYIAPEILFNSKEGHSFEVDLWSAGVVMYALLIGKPPF- 240 Query: 1328 QLNNVQAAHRIVADRHIEFDRDLYPNSGFDEILDTCLRSNQNERPNVFDL 1477 Q V+ +R + F ++ ++ +++ + L + + RP++ D+ Sbjct: 241 QDKEVKTIYRKIKANSYSFPSNVDISAEAKDLISSLLTHDPSIRPSIDDI 290
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 182,994,224
Number of Sequences: 369166
Number of extensions: 3922104
Number of successful extensions: 13101
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 11208
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 12035
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 18938402910
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail