DrC_00073
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
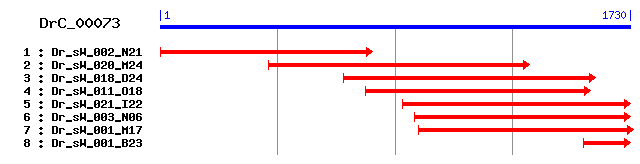
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00073 (1728 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P79384|PCCB_PIG Propionyl-CoA carboxylase beta chain, mi... 708 0.0 sp|P05166|PCCB_HUMAN Propionyl-CoA carboxylase beta chain, ... 705 0.0 sp|Q99MN9|PCCB_MOUSE Propionyl-CoA carboxylase beta chain, ... 704 0.0 sp|P07633|PCCB_RAT Propionyl-CoA carboxylase beta chain, mi... 696 0.0 sp|P53003|PCCB_SACER Propionyl-CoA carboxylase beta chain (... 454 e-127 sp|P96885|PCC5_MYCTU Probable propionyl-CoA carboxylase bet... 436 e-121 sp|P53002|PCCB_MYCLE Probable propionyl-CoA carboxylase bet... 434 e-121 sp|Q8GBW6|12S_PROFR Methylmalonyl-CoA carboxyltransferase 1... 415 e-115 sp|P54541|PCCB_BACSU Putative propionyl-CoA carboxylase bet... 408 e-113 sp|Q06101|PCCB_RHOER Propionyl-CoA carboxylase beta chain (... 243 2e-63
>sp|P79384|PCCB_PIG Propionyl-CoA carboxylase beta chain, mitochondrial precursor (PCCase beta subunit) (Propanoyl-CoA:carbon dioxide ligase beta subunit) Length = 539 Score = 708 bits (1827), Expect = 0.0 Identities = 357/512 (69%), Positives = 421/512 (82%), Gaps = 2/512 (0%) Frame = +2 Query: 107 LAVKKKIDETRNKALLGGGIKRIEAQHSKGKLTARERIHLLCDENEFVEWDAFMEHDCSD 286 ++V ++I+ R ALLGGG +RI++QH +GKLTARERI LL D F+E D F+EH C+D Sbjct: 33 VSVNERIENKRQAALLGGGQRRIDSQHKRGKLTARERISLLLDPGSFIESDMFVEHRCAD 92 Query: 287 FGMEA--NRFPGDSVVTGYGKIHGRPAFIFSQDFTVFGGSLSMAHAKKICKVMDQAMLVG 460 FGM A N+FPGDSVVTG G+I+GR ++FSQDFTVFGGSLS AHA+KICK+MDQAM VG Sbjct: 93 FGMAADKNKFPGDSVVTGRGRINGRLVYVFSQDFTVFGGSLSGAHAQKICKIMDQAMTVG 152 Query: 461 APVIWT**FWGCFVFKRVSHHFAGYAEIFQRNVMAFWCHSRKYL*LWGPLCGGVLFISPC 640 APVI G + + V AGYA+IF RNV A + L + GP GG ++ SP Sbjct: 153 APVIGLNDSGGARIQEGVES-LAGYADIFLRNVSASGVIPQISL-IMGPCAGGAVY-SPA 209 Query: 641 INPILLLWVKDSSYLFITGPDVVKSVTNEDVTQEELGGAKTHTTISGVAHKAFENDVDAL 820 + + VKD+SYLFITGPDVVKSVTNEDVTQEELGGA+THTT+SGVAH+AF+NDVDAL Sbjct: 210 LTDFTFM-VKDTSYLFITGPDVVKSVTNEDVTQEELGGARTHTTMSGVAHRAFDNDVDAL 268 Query: 821 MELREFFTFLPQSNRLHSPIRHSHDPPDRLVPSLDTIVPLESTAAYNMLDVIYGVVDESE 1000 LREFF +LP SN+ +PIR HDP DRLVP LDT+VPLEST AY+M+D+IY +VDE + Sbjct: 269 CNLREFFNYLPLSNQDPAPIRECHDPSDRLVPELDTVVPLESTRAYDMVDIIYSIVDERD 328 Query: 1001 FFEIMPTYAKNIIIGFGRMHGRTVGIIGNQPKVAAGCLDINASVKGARFVRFCDAFNIPL 1180 FFEIMP YAKNII+GF RM+GRTVGI+GNQPKVA+GCLDIN+SVKGARFVRFCDAFNIPL Sbjct: 329 FFEIMPNYAKNIIVGFARMNGRTVGIVGNQPKVASGCLDINSSVKGARFVRFCDAFNIPL 388 Query: 1181 LTFVDVPGFLPGTVQEYGGIIRHGAKLLYAYAEATVPKITIITRKAYGGAYDVMSSKHLR 1360 +TFVDVPGFLPGT QEYGGIIRHGAKLLYA+AEATVPKIT+ITRKAYGGAYDVMSSKHL Sbjct: 389 ITFVDVPGFLPGTAQEYGGIIRHGAKLLYAFAEATVPKITVITRKAYGGAYDVMSSKHLC 448 Query: 1361 GDINYAWPTAEVAVMGAKGAVQIIFRGSGSEQLTKEAEYVDKFANPFPAAVRGFVDDIIE 1540 GD NYAWPTAE+AVMGAKGAV+IIF+G + + +AEY++KFANPFPAAVRGFVDDII+ Sbjct: 449 GDTNYAWPTAEIAVMGAKGAVEIIFKGHENVE-AAQAEYIEKFANPFPAAVRGFVDDIIQ 507 Query: 1541 PRRTRAKICQDLDLLQHKQLINPWKKHGNIPL 1636 P TRA+IC DLD+L K++ PW+KH NIPL Sbjct: 508 PSSTRARICCDLDVLASKKVQRPWRKHANIPL 539
>sp|P05166|PCCB_HUMAN Propionyl-CoA carboxylase beta chain, mitochondrial precursor (PCCase beta subunit) (Propanoyl-CoA:carbon dioxide ligase beta subunit) Length = 539 Score = 705 bits (1819), Expect = 0.0 Identities = 358/511 (70%), Positives = 419/511 (81%), Gaps = 2/511 (0%) Frame = +2 Query: 110 AVKKKIDETRNKALLGGGIKRIEAQHSKGKLTARERIHLLCDENEFVEWDAFMEHDCSDF 289 +V ++I+ R ALLGGG +RI+AQH +GKLTARERI LL D FVE D F+EH C+DF Sbjct: 34 SVNERIENKRRTALLGGGQRRIDAQHKRGKLTARERISLLLDPGSFVESDMFVEHRCADF 93 Query: 290 GMEA--NRFPGDSVVTGYGKIHGRPAFIFSQDFTVFGGSLSMAHAKKICKVMDQAMLVGA 463 GM A N+FPGDSVVTG G+I+GR ++FSQDFTVFGGSLS AHA+KICK+MDQA+ VGA Sbjct: 94 GMAADKNKFPGDSVVTGRGRINGRLVYVFSQDFTVFGGSLSGAHAQKICKIMDQAITVGA 153 Query: 464 PVIWT**FWGCFVFKRVSHHFAGYAEIFQRNVMAFWCHSRKYL*LWGPLCGGVLFISPCI 643 PVI G + + V AGYA+IF RNV A + L + GP GG ++ SP + Sbjct: 154 PVIGLNDSGGARIQEGVES-LAGYADIFLRNVTASGVIPQISL-IMGPCAGGAVY-SPAL 210 Query: 644 NPILLLWVKDSSYLFITGPDVVKSVTNEDVTQEELGGAKTHTTISGVAHKAFENDVDALM 823 + VKD+SYLFITGPDVVKSVTNEDVTQEELGGAKTHTT+SGVAH+AFENDVDAL Sbjct: 211 TDFTFM-VKDTSYLFITGPDVVKSVTNEDVTQEELGGAKTHTTMSGVAHRAFENDVDALC 269 Query: 824 ELREFFTFLPQSNRLHSPIRHSHDPPDRLVPSLDTIVPLESTAAYNMLDVIYGVVDESEF 1003 LR+FF +LP S++ + +R HDP DRLVP LDTIVPLEST AYNM+D+I+ VVDE EF Sbjct: 270 NLRDFFNYLPLSSQDPASVRECHDPSDRLVPELDTIVPLESTKAYNMVDIIHSVVDEREF 329 Query: 1004 FEIMPTYAKNIIIGFGRMHGRTVGIIGNQPKVAAGCLDINASVKGARFVRFCDAFNIPLL 1183 FEIMP YAKNII+GF RM+GRTVGI+GNQPKVA+GCLDIN+SVKGARFVRFCDAFNIPL+ Sbjct: 330 FEIMPNYAKNIIVGFARMNGRTVGIVGNQPKVASGCLDINSSVKGARFVRFCDAFNIPLI 389 Query: 1184 TFVDVPGFLPGTVQEYGGIIRHGAKLLYAYAEATVPKITIITRKAYGGAYDVMSSKHLRG 1363 TFVDVPGFLPGT QEYGGIIRHGAKLLYA+AEATVPK+T+ITRKAYGGAYDVMSSKHL G Sbjct: 390 TFVDVPGFLPGTAQEYGGIIRHGAKLLYAFAEATVPKVTVITRKAYGGAYDVMSSKHLCG 449 Query: 1364 DINYAWPTAEVAVMGAKGAVQIIFRGSGSEQLTKEAEYVDKFANPFPAAVRGFVDDIIEP 1543 D NYAWPTAE+AVMGAKGAV+IIF+G + + +AEY++KFANPFPAAVRGFVDDII+P Sbjct: 450 DTNYAWPTAEIAVMGAKGAVEIIFKGHENVE-AAQAEYIEKFANPFPAAVRGFVDDIIQP 508 Query: 1544 RRTRAKICQDLDLLQHKQLINPWKKHGNIPL 1636 TRA+IC DLD+L K++ PW+KH NIPL Sbjct: 509 SSTRARICCDLDVLASKKVQRPWRKHANIPL 539
>sp|Q99MN9|PCCB_MOUSE Propionyl-CoA carboxylase beta chain, mitochondrial precursor (PCCase beta subunit) (Propanoyl-CoA:carbon dioxide ligase beta subunit) Length = 541 Score = 704 bits (1817), Expect = 0.0 Identities = 356/512 (69%), Positives = 421/512 (82%), Gaps = 2/512 (0%) Frame = +2 Query: 107 LAVKKKIDETRNKALLGGGIKRIEAQHSKGKLTARERIHLLCDENEFVEWDAFMEHDCSD 286 ++VK++ID R+ ALLGGG +RI+AQH +GKLTARERI LL D F+E D F+EH C+D Sbjct: 35 VSVKERIDNKRHAALLGGGQRRIDAQHKRGKLTARERISLLLDPGSFMESDMFVEHRCAD 94 Query: 287 FGMEA--NRFPGDSVVTGYGKIHGRPAFIFSQDFTVFGGSLSMAHAKKICKVMDQAMLVG 460 FGM A N+FPGDSVVTG G+I+GR ++FSQDFTVFGGSLS AHA+KICK+MDQA+ VG Sbjct: 95 FGMAADKNKFPGDSVVTGRGRINGRLVYVFSQDFTVFGGSLSGAHAQKICKIMDQAITVG 154 Query: 461 APVIWT**FWGCFVFKRVSHHFAGYAEIFQRNVMAFWCHSRKYL*LWGPLCGGVLFISPC 640 APVI G + + V AGYA+IF RNV A + L + GP GG ++ SP Sbjct: 155 APVIGLNDSGGARIQEGVES-LAGYADIFLRNVTASGVIPQISL-IMGPCAGGAVY-SPA 211 Query: 641 INPILLLWVKDSSYLFITGPDVVKSVTNEDVTQEELGGAKTHTTISGVAHKAFENDVDAL 820 + + VKD+SYLFITGP+VVKSVTNEDVTQE+LGGAKTHTT+SGVAH+AF+NDVDAL Sbjct: 212 LTDFTFM-VKDTSYLFITGPEVVKSVTNEDVTQEQLGGAKTHTTVSGVAHRAFDNDVDAL 270 Query: 821 MELREFFTFLPQSNRLHSPIRHSHDPPDRLVPSLDTIVPLESTAAYNMLDVIYGVVDESE 1000 LREFF FLP S++ +PIR HDP DRLVP LDT+VPLES+ AYNMLD+I+ V+DE E Sbjct: 271 CNLREFFNFLPLSSQDPAPIRECHDPSDRLVPELDTVVPLESSKAYNMLDIIHAVIDERE 330 Query: 1001 FFEIMPTYAKNIIIGFGRMHGRTVGIIGNQPKVAAGCLDINASVKGARFVRFCDAFNIPL 1180 FFEIMP+YA NI++GF RM+GRTVGI+GNQP VA+GCLDIN+SVKGARFVRFCDAFNIPL Sbjct: 331 FFEIMPSYALNIVVGFARMNGRTVGIVGNQPNVASGCLDINSSVKGARFVRFCDAFNIPL 390 Query: 1181 LTFVDVPGFLPGTVQEYGGIIRHGAKLLYAYAEATVPKITIITRKAYGGAYDVMSSKHLR 1360 +TFVDVPGFLPGT QEYGGIIRHGAKLLYA+AEATVPKIT+ITRKAYGGAYDVMSSKHL Sbjct: 391 ITFVDVPGFLPGTAQEYGGIIRHGAKLLYAFAEATVPKITVITRKAYGGAYDVMSSKHLL 450 Query: 1361 GDINYAWPTAEVAVMGAKGAVQIIFRGSGSEQLTKEAEYVDKFANPFPAAVRGFVDDIIE 1540 GD NYAWPTAE+AVMGAKGAV+IIF+G + +AEYV+KFANPFPAAVRGFVDDII+ Sbjct: 451 GDTNYAWPTAEIAVMGAKGAVEIIFKGHQDVE-AAQAEYVEKFANPFPAAVRGFVDDIIQ 509 Query: 1541 PRRTRAKICQDLDLLQHKQLINPWKKHGNIPL 1636 P TRA+IC DL++L K++ PW+KH NIPL Sbjct: 510 PSSTRARICCDLEVLASKKVHRPWRKHANIPL 541
>sp|P07633|PCCB_RAT Propionyl-CoA carboxylase beta chain, mitochondrial precursor (PCCase beta subunit) (Propanoyl-CoA:carbon dioxide ligase beta subunit) Length = 541 Score = 696 bits (1796), Expect = 0.0 Identities = 352/512 (68%), Positives = 417/512 (81%), Gaps = 2/512 (0%) Frame = +2 Query: 107 LAVKKKIDETRNKALLGGGIKRIEAQHSKGKLTARERIHLLCDENEFVEWDAFMEHDCSD 286 ++V ++I+ R+ ALLGGG +RI+AQH +GKLTARERI LL D F+E D F+EH C+D Sbjct: 35 VSVNERIENKRHAALLGGGQRRIDAQHKRGKLTARERISLLLDPGSFLESDMFVEHRCAD 94 Query: 287 FGM--EANRFPGDSVVTGYGKIHGRPAFIFSQDFTVFGGSLSMAHAKKICKVMDQAMLVG 460 FGM E N+FPGDSVVTG G+I+GR ++FSQDFTVFGGSLS AHA+KICK+MDQA+ VG Sbjct: 95 FGMAAEKNKFPGDSVVTGRGRINGRLVYVFSQDFTVFGGSLSGAHAQKICKIMDQAITVG 154 Query: 461 APVIWT**FWGCFVFKRVSHHFAGYAEIFQRNVMAFWCHSRKYL*LWGPLCGGVLFISPC 640 APVI G + + V AGYA+IF RNV A + L + GP GG ++ SP Sbjct: 155 APVIGLNDSGGARIQEGVES-LAGYADIFLRNVTASGVIPQISL-IMGPCAGGAVY-SPA 211 Query: 641 INPILLLWVKDSSYLFITGPDVVKSVTNEDVTQEELGGAKTHTTISGVAHKAFENDVDAL 820 + + VKD+SYLFITGP+ VKSVTNEDVTQE+LGGAKTHTT+SGVAH+AF+NDVDAL Sbjct: 212 LTDFTFM-VKDTSYLFITGPEFVKSVTNEDVTQEQLGGAKTHTTVSGVAHRAFDNDVDAL 270 Query: 821 MELREFFTFLPQSNRLHSPIRHSHDPPDRLVPSLDTIVPLESTAAYNMLDVIYGVVDESE 1000 LREF FLP SN+ + IR HDP DRLVP LDT+VPLES+ AYNMLD+I+ V+DE E Sbjct: 271 CNLREFLNFLPLSNQDPASIRECHDPSDRLVPELDTVVPLESSKAYNMLDIIHAVIDERE 330 Query: 1001 FFEIMPTYAKNIIIGFGRMHGRTVGIIGNQPKVAAGCLDINASVKGARFVRFCDAFNIPL 1180 FFEIMP YAKNI+IGF RM+GRTVGI+GNQP VA+GCLDIN+SVKGARFVRFCDAF+IPL Sbjct: 331 FFEIMPNYAKNIVIGFARMNGRTVGIVGNQPNVASGCLDINSSVKGARFVRFCDAFSIPL 390 Query: 1181 LTFVDVPGFLPGTVQEYGGIIRHGAKLLYAYAEATVPKITIITRKAYGGAYDVMSSKHLR 1360 +TFVDVPGFLPGT QEYGGIIRHGAKLLYA+AEATVPKIT+ITRKAYGGAYDVMSSKHL Sbjct: 391 ITFVDVPGFLPGTAQEYGGIIRHGAKLLYAFAEATVPKITVITRKAYGGAYDVMSSKHLL 450 Query: 1361 GDINYAWPTAEVAVMGAKGAVQIIFRGSGSEQLTKEAEYVDKFANPFPAAVRGFVDDIIE 1540 GD NYAWPTAE+AVMGAKGAV+IIF+G + +AEYV+KFANPFPAAVRGFVDDII+ Sbjct: 451 GDTNYAWPTAEIAVMGAKGAVEIIFKGHEDVE-AAQAEYVEKFANPFPAAVRGFVDDIIQ 509 Query: 1541 PRRTRAKICQDLDLLQHKQLINPWKKHGNIPL 1636 P TRA+IC DL++L K++ PW+KH N+PL Sbjct: 510 PSSTRARICCDLEVLASKKVHRPWRKHANVPL 541
>sp|P53003|PCCB_SACER Propionyl-CoA carboxylase beta chain (PCCase) (Propanoyl-CoA:carbon dioxide ligase) Length = 546 Score = 454 bits (1168), Expect = e-127 Identities = 257/532 (48%), Positives = 334/532 (62%), Gaps = 20/532 (3%) Frame = +2 Query: 101 HTLAVKKKIDETRN-KALLGGGIKRIEAQHSKGKLTARERIHLLCDENEFVEWDAFMEHD 277 HT A K RN +A+ G + + QH+KGK TARERI +L DE FVE D H Sbjct: 19 HTTAGKLADLYRRNHEAVHAGSERAVAKQHAKGKRTARERIDMLLDEGSFVELDEHARHR 78 Query: 278 CSDFGMEANRFPGDSVVTGYGKIHGRPAFIFSQDFTVFGGSLSMAHAKKICKVMDQAMLV 457 ++FGM+A+R GD VVTG+G + GR +FSQDFTVFGGSL +KI KVMD AM Sbjct: 79 STNFGMDADRPYGDGVVTGWGTVDGRRVCVFSQDFTVFGGSLGEVFGEKIVKVMDLAMKT 138 Query: 458 GAPVIWT**FWGCFVFKRVSHHFAGYAEIFQRNVMAFWCHSRKYL*LWGPLCGGVLFISP 637 G P++ G + + V+ YAEIF+RN A + L + GP GG ++ SP Sbjct: 139 GCPLVGINDSGGARIQEGVAA-LGLYAEIFKRNTHASGVIPQISL-IMGPCAGGAVY-SP 195 Query: 638 CINPILLLWVKDSSYLFITGPDVVKSVTNEDVTQEELGGAKTHTTISGVAHKAFENDVDA 817 I ++ V +S++FITGPDV+K+VT EDV+ E+LGGA+TH SG AH ++ DA Sbjct: 196 AITDFTVM-VDQTSHMFITGPDVIKTVTGEDVSFEDLGGARTHNERSGNAHYLATDEDDA 254 Query: 818 LMELREFFTFLPQSNRLHSPIRHSHDPPDRLVPS--------LDTIVPLESTAAYNMLDV 973 + ++E +FLP +N SP+ + + V LD +VP Y+M +V Sbjct: 255 ISYVKELLSFLPSNNLSSSPVFPGAEVEEGSVADGVGDADLELDALVPDSPNQPYDMREV 314 Query: 974 IYGVVDESEFFEIMPTYAKNIIIGFGRMHGRTVGIIGNQPKVAAGCLDINASVKGARFVR 1153 I +VDE EF E+ +A N++ GFGR+ G +VG++ NQP AG LDI+AS K ARFVR Sbjct: 315 ITRLVDEGEFLEVSALFAPNMLCGFGRIEGASVGVVANQPMQLAGTLDIDASEKAARFVR 374 Query: 1154 FCDAFNIPLLTFVDVPGFLPGTVQEYGGIIRHGAKLLYAYAEATVPKITIITRKAYGGAY 1333 FCDAFNIP+LT VDVPGFLPGT Q++ IIR GAKLLYAYAEATVP +T+ITRKAYGGAY Sbjct: 375 FCDAFNIPVLTLVDVPGFLPGTGQDWNAIIRRGAKLLYAYAEATVPLVTVITRKAYGGAY 434 Query: 1334 DVMSSKHLRGDINYAWPTAEVAVMGAKGAVQIIFRGSGSEQLTK-----------EAEYV 1480 DVM SKHL DIN AWPTA++AVMGA+GA I++R +E + + EY Sbjct: 435 DVMGSKHLGADINLAWPTAQIAVMGAQGAANILYRRQLAEAAERGEDVEALRARLQQEYE 494 Query: 1481 DKFANPFPAAVRGFVDDIIEPRRTRAKICQDLDLLQHKQLINPWKKHGNIPL 1636 D NP+ AA RG+VD +I P TR + + L +L K+ P KKHGNIPL Sbjct: 495 DTLCNPYVAAERGYVDSVIPPSHTRGHVARALRMLADKREALPAKKHGNIPL 546
>sp|P96885|PCC5_MYCTU Probable propionyl-CoA carboxylase beta chain 5 (PCCase) (Propanoyl-CoA:carbon dioxide ligase) Length = 548 Score = 436 bits (1120), Expect = e-121 Identities = 254/531 (47%), Positives = 329/531 (61%), Gaps = 19/531 (3%) Frame = +2 Query: 101 HTLAVK-KKIDETRNKALLGGGIKRIEAQHSKGKLTARERIHLLCDENEFVEWDAFMEHD 277 HT A K ++ + R ++L G +E H+KGKLTARERI+ L DE+ FVE DA +H Sbjct: 23 HTTAGKLAELHKRREESLHPVGEDAVEKVHAKGKLTARERIYALLDEDSFVELDALAKHR 82 Query: 278 CSDFGMEANRFPGDSVVTGYGKIHGRPAFIFSQDFTVFGGSLSMAHAKKICKVMDQAMLV 457 ++F + R GD VVTGYG I GR IFSQD TVFGGSL + +KI KV + A+ Sbjct: 83 STNFNLGEKRPLGDGVVTGYGTIDGRDVCIFSQDATVFGGSLGEVYGEKIVKVQELAIKT 142 Query: 458 GAPVIWT**FWGCFVFKRVSHHFAGYAEIFQRNVMAFWCHSRKYL*LWGPLCGGVLFISP 637 G P+I G + + V Y+ IF+ N++A + L + G GG ++ SP Sbjct: 143 GRPLIGINDGAGARIQEGVVS-LGLYSRIFRNNILASGVIPQISL-IMGAAAGGHVY-SP 199 Query: 638 CINPILLLWVKDSSYLFITGPDVVKSVTNEDVTQEELGGAKTHTTISGVAHKAFENDVDA 817 + +++ V +S +FITGPDV+K+VT E+VT EELGGA TH SG AH A + DA Sbjct: 200 ALTDFVIM-VDQTSQMFITGPDVIKTVTGEEVTMEELGGAHTHMAKSGTAHYAASGEQDA 258 Query: 818 LMELREFFTFLPQSNRLHSPIRHSHDPPDRLVPSL-------DTIVPLESTAAYNMLDVI 976 +RE ++LP +N +P + P + +L DT++P Y+M +VI Sbjct: 259 FDYVRELLSYLPPNNSTDAPRYQAAAPTGPIEENLTDEDLELDTLIPDSPNQPYDMHEVI 318 Query: 977 YGVVDESEFFEIMPTYAKNIIIGFGRMHGRTVGIIGNQPKVAAGCLDINASVKGARFVRF 1156 ++D+ EF EI YA+NI++GFGR+ GR VGI+ NQP AGCLDINAS K ARFVR Sbjct: 319 TRLLDD-EFLEIQAGYAQNIVVGFGRIDGRPVGIVANQPTHFAGCLDINASEKAARFVRT 377 Query: 1157 CDAFNIPLLTFVDVPGFLPGTVQEYGGIIRHGAKLLYAYAEATVPKITIITRKAYGGAYD 1336 CD FNIP++ VDVPGFLPGT QEY GIIR GAKLLYAY EATVPKIT+ITRKAYGGAY Sbjct: 378 CDCFNIPIVMLVDVPGFLPGTDQEYNGIIRRGAKLLYAYGEATVPKITVITRKAYGGAYC 437 Query: 1337 VMSSKHLRGDINYAWPTAEVAVMGAKGAVQIIFRGSGSE-----------QLTKEAEYVD 1483 VM SK + D+N AWPTA++AVMGA GAV ++R +E +L + EY D Sbjct: 438 VMGSKDMGCDVNLAWPTAQIAVMGASGAVGFVYRQQLAEAAANGEDIDKLRLRLQQEYED 497 Query: 1484 KFANPFPAAVRGFVDDIIEPRRTRAKICQDLDLLQHKQLINPWKKHGNIPL 1636 NP+ AA RG+VD +I P TR I L LL+ K P KKHGN+PL Sbjct: 498 TLVNPYVAAERGYVDAVIPPSHTRGYIGTALRLLERKIAQLPPKKHGNVPL 548
>sp|P53002|PCCB_MYCLE Probable propionyl-CoA carboxylase beta chain 5 (PCCase) (Propanoyl-CoA:carbon dioxide ligase) Length = 549 Score = 434 bits (1116), Expect = e-121 Identities = 253/532 (47%), Positives = 326/532 (61%), Gaps = 20/532 (3%) Frame = +2 Query: 101 HTLAVK-KKIDETRNKALLGGGIKRIEAQHSKGKLTARERIHLLCDENEFVEWDAFMEHD 277 HT A K ++ + +AL G E H+KGK TARERI+ L D++ FVE DA H Sbjct: 23 HTTAGKLAELHKRTEEALHPVGAAAFEKVHAKGKFTARERIYALLDDDSFVELDALARHR 82 Query: 278 CSDFGMEANRFPGDSVVTGYGKIHGRPAFIFSQDFTVFGGSLSMAHAKKICKVMDQAMLV 457 ++FG+ NR GD VVTGYG I GR IFSQD TVFGGSL + +KI KV + A+ Sbjct: 83 STNFGLGENRPVGDGVVTGYGTIDGRDVCIFSQDVTVFGGSLGEVYGEKIVKVQELAIKT 142 Query: 458 GAPVIWT**FWGCFVFKRVSHHFAGYAEIFQRNVMAFWCHSRKYL*LWGPLCGGVLFISP 637 G P+I G + + V Y+ IF+ N++A + L + G GG ++ SP Sbjct: 143 GRPLIGINDGAGARIQEGVVS-LGLYSRIFRNNILASGVIPQISL-IMGAAAGGHVY-SP 199 Query: 638 CINPILLLWVKDSSYLFITGPDVVKSVTNEDVTQEELGGAKTHTTISGVAHKAFENDVDA 817 + +++ V +S +FITGPDV+K+VT EDVT EELGGA TH SG AH + DA Sbjct: 200 ALTDFVVM-VDQTSQMFITGPDVIKTVTGEDVTMEELGGAHTHMAKSGTAHYVASGEQDA 258 Query: 818 LMELREFFTFLPQSNRLHSPIRHSHDPP-----DRLVPS---LDTIVPLESTAAYNMLDV 973 +R+ ++LP +N +P R+S P D L LDT++P Y+M +V Sbjct: 259 FDWVRDVLSYLPSNNFTDAP-RYSKPVPHGSIEDNLTAKDLELDTLIPDSPNQPYDMHEV 317 Query: 974 IYGVVDESEFFEIMPTYAKNIIIGFGRMHGRTVGIIGNQPKVAAGCLDINASVKGARFVR 1153 + ++DE EF E+ YA NI++G GR+ R VGI+ NQP AGCLDINAS K ARFVR Sbjct: 318 VTRLLDEEEFLEVQAGYATNIVVGLGRIDDRPVGIVANQPIQFAGCLDINASEKAARFVR 377 Query: 1154 FCDAFNIPLLTFVDVPGFLPGTVQEYGGIIRHGAKLLYAYAEATVPKITIITRKAYGGAY 1333 CD FNIP++ VDVPGFLPGT QEY GIIR GAKLL+AY EATVPKIT+ITRKAYGGAY Sbjct: 378 VCDCFNIPIVMLVDVPGFLPGTEQEYDGIIRRGAKLLFAYGEATVPKITVITRKAYGGAY 437 Query: 1334 DVMSSKHLRGDINYAWPTAEVAVMGAKGAVQIIFRGSGSE-----------QLTKEAEYV 1480 VM SK++ D+N AWPTA++AVMGA GAV ++R ++ +L + EY Sbjct: 438 CVMGSKNMGCDVNLAWPTAQIAVMGASGAVGFVYRKELAQAAKNGANVDELRLQLQQEYE 497 Query: 1481 DKFANPFPAAVRGFVDDIIEPRRTRAKICQDLDLLQHKQLINPWKKHGNIPL 1636 D NP+ AA RG+VD +I P TR I L LL+ K P KKHGNIPL Sbjct: 498 DTLVNPYIAAERGYVDAVIPPSHTRGYIATALHLLERKIAHLPPKKHGNIPL 549
>sp|Q8GBW6|12S_PROFR Methylmalonyl-CoA carboxyltransferase 12S subunit (Transcarboxylase 12S subunit) Length = 611 Score = 415 bits (1066), Expect = e-115 Identities = 233/529 (44%), Positives = 325/529 (61%), Gaps = 9/529 (1%) Frame = +2 Query: 77 AAPLQGSAHTLAVKKKIDETRNKALLGGGIKRIEAQHSKGKLTARERIHLLCDENEFVEW 256 A+ ++G LA ++++ E GGG +R+E QHS+GK TARER++ L D + F E Sbjct: 10 ASTMEGRVEQLAEQRQVIEA------GGGERRVEKQHSQGKQTARERLNNLLDPHSFDEV 63 Query: 257 DAFMEHDCSDFGMEANRFPGDSVVTGYGKIHGRPAFIFSQDFTVFGGSLSMAHAKKICKV 436 AF +H + FGM+ P D VVTG G I GRP SQDFTV GGS + K+ + Sbjct: 64 GAFRKHRTTLFGMDKAVVPADGVVTGRGTILGRPVHAASQDFTVMGGSAGETQSTKVVET 123 Query: 437 MDQAMLVGAPVIWT**FWGCFVFKRVSHHFAGYAEIFQRNVMAFWCHSRKYL*LWGPLCG 616 M+QA+L G P ++ G + + + +GY ++F NV + + + GP G Sbjct: 124 MEQALLTGTPFLFFYDSGGARIQEGIDS-LSGYGKMFFANVKLSGVVPQIAI-IAGPCAG 181 Query: 617 GVLFISPCINPILLLWVKDSSYLFITGPDVVKSVTNEDVTQEELGGAKTHTTISGVAHKA 796 G + SP + +++ K +++FITGP V+KSVT EDVT +ELGGA+ H ISG H Sbjct: 182 GASY-SPALTDFIIMTKK--AHMFITGPQVIKSVTGEDVTADELGGAEAHMAISGNIHFV 238 Query: 797 FENDVDALMELREFFTFLPQSNRLHSPIRHSHDPPDRLVPS--LDTIVPLESTAAYNMLD 970 E+D A + ++ +FLPQ+N + + P + + P+ L IVP++ Y++ D Sbjct: 239 AEDDDAAELIAKKLLSFLPQNNTEEASFVN---PNNDVSPNTELRDIVPIDGKKGYDVRD 295 Query: 971 VIYGVVDESEFFEIMPTYAKNIIIGFGRMHGRTVGIIGNQPKVAAGCLDINASVKGARFV 1150 VI +VD ++ E+ YA N++ F R++GR+VGI+ NQP V +GCLDINAS K A FV Sbjct: 296 VIAKIVDWGDYLEVKAGYATNLVTAFARVNGRSVGIVANQPSVMSGCLDINASDKAAEFV 355 Query: 1151 RFCDAFNIPLLTFVDVPGFLPGTVQEYGGIIRHGAKLLYAYAEATVPKITIITRKAYGGA 1330 FCD+FNIPL+ VDVPGFLPG QEYGGIIRHGAK+LYAY+EATVPKIT++ RKAYGG+ Sbjct: 356 NFCDSFNIPLVQLVDVPGFLPGVQQEYGGIIRHGAKMLYAYSEATVPKITVVLRKAYGGS 415 Query: 1331 YDVMSSKHLRGDINYAWPTAEVAVMGAKGAVQIIFR-------GSGSEQLTKEAEYVDKF 1489 Y M ++ L D YAWP+AE+AVMGA+GA +IFR + + K EY + F Sbjct: 416 YLAMCNRDLGADAVYAWPSAEIAVMGAEGAANVIFRKEIKAADDPDAMRAEKIEEYQNAF 475 Query: 1490 ANPFPAAVRGFVDDIIEPRRTRAKICQDLDLLQHKQLINPWKKHGNIPL 1636 P+ AA RG VDD+I+P TR KI L++ K+ P KK +PL Sbjct: 476 NTPYVAAARGQVDDVIDPADTRRKIASALEMYATKRQTRPAKKPWKLPL 524
>sp|P54541|PCCB_BACSU Putative propionyl-CoA carboxylase beta chain (PCCase) (Propanoyl-CoA:carbon dioxide ligase) Length = 506 Score = 408 bits (1048), Expect = e-113 Identities = 233/509 (45%), Positives = 311/509 (61%), Gaps = 9/509 (1%) Frame = +2 Query: 137 RNKALLGGGIKRIEAQHSKGKLTARERIHLLCDENEFVEWDAFMEHDCSDFGMEANRFPG 316 R +A GGG +++ Q KGKLTARERI L D++ F+E FME S R G Sbjct: 11 RKQAEEGGGREKLAQQRQKGKLTARERIIFLLDQDSFIELHPFME---SQVLTREQRMLG 67 Query: 317 DSVVTGYGKIHGRPAFIFSQDFTVFGGSLSMAHAKKICKVMDQAMLVGAPVIWT**FWGC 496 D VVTGYG I GR ++F+QDFTV+GG+L HA+KIC +MD A AP+I G Sbjct: 68 DGVVTGYGTIDGRSVYVFAQDFTVYGGALGETHARKICALMDLAAKNKAPIIGLNDSGGA 127 Query: 497 FVFKRVSHHFAGYAEIFQRNVMAFWCHSRKYL*LWGPLCGGVLFISPCINPILLLWVKDS 676 + + V GY IF RNV+ + + L GP GG ++ SP + + + + + Sbjct: 128 RIQEGVLS-LDGYGHIFYRNVLYSGVIPQISVIL-GPCAGGAVY-SPALTDFIFM-AEQT 183 Query: 677 SYLFITGPDVVKSVTNEDVTQEELGGAKTHTTISGVAHKAFENDVDALMELREFFTFLPQ 856 +FITGP V++ VT E V E LGGA H +SG AH + + + L +R+ ++LP Sbjct: 184 GRMFITGPKVIEKVTGEQVDAESLGGAGIHNAVSGNAHFSGHTEKEVLTGVRKLLSYLPL 243 Query: 857 SNRLHSPIRHSHDPPDRLV--PSLDTIVPLESTAAYNMLDVIYGVVDESEFFEIMPTYAK 1030 + R P P++ P L+ +VP ++T Y++ VI + D FFEI P +AK Sbjct: 244 NGRTTEP------KPEKEASRPLLNRLVPADTTKPYDVRKVIRELADPQSFFEIQPFFAK 297 Query: 1031 NIIIGFGRMHGRTVGIIGNQPKVAAGCLDINASVKGARFVRFCDAFNIPLLTFVDVPGFL 1210 NI+IGF R+ + +GI+ +QPK AG L I+A+ K ARF+RFCDAF+IPLLT DVPGFL Sbjct: 298 NIVIGFARLGEKAIGIVASQPKHLAGSLTIDAADKAARFIRFCDAFDIPLLTVEDVPGFL 357 Query: 1211 PGTVQEYGGIIRHGAKLLYAYAEATVPKITIITRKAYGGAYDVMSSKHLRGDINYAWPTA 1390 PG QE+ GIIRHGAKLL+AYAEATVPK+T+I RKAYGGAY M+SK + D+ +AWP A Sbjct: 358 PGIQQEHNGIIRHGAKLLFAYAEATVPKVTLIIRKAYGGAYVAMNSKAIGADLVFAWPNA 417 Query: 1391 EVAVMGAKGAVQIIF----RGSGSEQLTKE---AEYVDKFANPFPAAVRGFVDDIIEPRR 1549 E+AVMG +GA I++ + S Q TK AEY + A P+ AA G VDDII P Sbjct: 418 EIAVMGPEGAASILYEKEIKASADPQKTKREKTAEYKKQNAGPYKAAACGMVDDIILPEE 477 Query: 1550 TRAKICQDLDLLQHKQLINPWKKHGNIPL 1636 +R ++ Q +L HK P KKHGNIPL Sbjct: 478 SRGRLIQAFHMLTHKTEERPKKKHGNIPL 506
>sp|Q06101|PCCB_RHOER Propionyl-CoA carboxylase beta chain (PCCase) (Propanoyl-CoA:carbon dioxide ligase) Length = 476 Score = 243 bits (619), Expect = 2e-63 Identities = 152/416 (36%), Positives = 225/416 (54%), Gaps = 5/416 (1%) Frame = +2 Query: 323 VVTGYGKIHGRPAFIFSQDFTVFGGSLSMAHAKKICKVMDQAMLVGAPVIWT**FWGCFV 502 V+ G I G + D TV GG++ + K + K +D A+ +P++ G + Sbjct: 43 VLAASGNIDGVRTIAYCSDATVMGGAMGVDGCKHLVKAIDTAIEEESPIVGLWHSGGARL 102 Query: 503 FKRVSH-HFAGYAEIFQRNVMAFWCHSRKYL*LWGPLCGGVLFISPCINPILLLWVKDSS 679 + V H G +F+ V A + + L G GG + P + ++++ Sbjct: 103 AEGVEALHAVGL--VFEAMVRASGLIPQISVVL-GFAAGGAAY-GPALTDVVIM--APEG 156 Query: 680 YLFITGPDVVKSVTNEDVTQEELGGAKTHTTISGVAHKAFENDVDALMELREFFTFLPQS 859 +F+TGPDVV+SVT E V LGG THT SGVAH A ++ DAL R + + + Sbjct: 157 RVFVTGPDVVRSVTGEQVDMVSLGGPDTHTKKSGVAHIAAHDEADALHRARRLVSMMCEQ 216 Query: 860 NRLHSPIRHSHDPPDRLVPSLDTIVPLESTAAYNMLDVIYGVVD----ESEFFEIMPTYA 1027 D L ++P + AY++ +++ ++D ES F E+ YA Sbjct: 217 GEFDQRAAELGDS------DLRAMMPASAKRAYDVRPIVHEMLDNVEGESSFEELQGNYA 270 Query: 1028 KNIIIGFGRMHGRTVGIIGNQPKVAAGCLDINASVKGARFVRFCDAFNIPLLTFVDVPGF 1207 ++I+ GFGRM GRTVG+I N P + GCL+ ++ K ARFVR C+AF +PL+ VDVPG+ Sbjct: 271 RSIVTGFGRMAGRTVGVIANNP-LRLGCLNSESAEKAARFVRLCNAFGVPLVVVVDVPGY 329 Query: 1208 LPGTVQEYGGIIRHGAKLLYAYAEATVPKITIITRKAYGGAYDVMSSKHLRGDINYAWPT 1387 LPG E+ G++R GAKLL+A+AEATVP++T++TRK YGGAY M+S+ L +AWP Sbjct: 330 LPGVSMEWEGVVRRGAKLLHAFAEATVPRVTVVTRKIYGGAYIAMNSRALGATAVFAWPN 389 Query: 1388 AEVAVMGAKGAVQIIFRGSGSEQLTKEAEYVDKFANPFPAAVRGFVDDIIEPRRTR 1555 +EVAVMGAK AV I+ + + + E E + A+ G VD RR R Sbjct: 390 SEVAVMGAKAAVGILHKRALAAAPDDEREALHDRLAAEHEAIAGGVDRRCRNRRRR 445
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 212,316,853
Number of Sequences: 369166
Number of extensions: 4803462
Number of successful extensions: 11962
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 11036
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 11867
length of database: 68,354,980
effective HSP length: 116
effective length of database: 46,925,720
effective search space used: 21538905480
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail