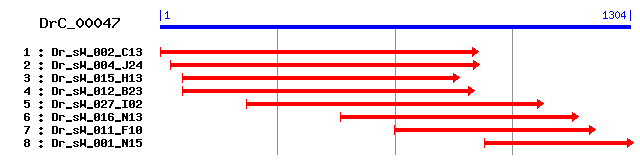
DrC_00047
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00047 (1304 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P05091|ALDH2_HUMAN Aldehyde dehydrogenase, mitochondrial... 548 e-155 sp|P81178|ALDH2_MESAU Aldehyde dehydrogenase, mitochondrial... 545 e-155 sp|P11884|ALDH2_RAT Aldehyde dehydrogenase, mitochondrial p... 545 e-154 sp|P47738|ALDH2_MOUSE Aldehyde dehydrogenase, mitochondrial... 543 e-154 sp|P20000|ALDH2_BOVIN Aldehyde dehydrogenase, mitochondrial... 540 e-153 sp|P12762|ALDH2_HORSE Aldehyde dehydrogenase, mitochondrial... 532 e-151 sp|O94788|AL1A2_HUMAN Retinal dehydrogenase 2 (RalDH2) (RAL... 523 e-148 sp|O93344|AL1A2_CHICK Retinal dehydrogenase 2 (RalDH2) (RAL... 522 e-148 sp|Q63639|AL1A2_RAT Retinal dehydrogenase 2 (RalDH2) (RALDH... 521 e-147 sp|Q62148|AL1A2_MOUSE Retinal dehydrogenase 2 (RalDH2) (RAL... 520 e-147
>sp|P05091|ALDH2_HUMAN Aldehyde dehydrogenase, mitochondrial precursor (ALDH class 2) (ALDHI) (ALDH-E2) Length = 517 Score = 548 bits (1412), Expect = e-155 Identities = 255/373 (68%), Positives = 306/373 (82%), Gaps = 1/373 (0%) Frame = +1 Query: 19 CYEYYAGWADKNHGKVIPMRGNYLGFTRHEAVGICGQIIPWNFPLLMQAWKLGPALCMGN 198 C YYAGWADK HGK IP+ G++ +TRHE VG+CGQIIPWNFPLLMQAWKLGPAL GN Sbjct: 145 CLRYYAGWADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQAWKLGPALATGN 204 Query: 199 TVVMKTAEQTPLTANYVASLIAEAGFPPGVVNIVPGMGPVAGAALVSHPNVDKIAFTGST 378 VVMK AEQTPLTA YVA+LI EAGFPPGVVNIVPG GP AGAA+ SH +VDK+AFTGST Sbjct: 205 VVVMKVAEQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASHEDVDKVAFTGST 264 Query: 379 EVGRIISSECNKNNIKRLTLELGGKSPNIVLADADFDHAVKSSHFALFFNMGQCCCAGSR 558 E+GR+I +N+KR+TLELGGKSPNI+++DAD D AV+ +HFALFFN GQCCCAGSR Sbjct: 265 EIGRVIQVAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALFFNQGQCCCAGSR 324 Query: 559 IYVEEKIYDKFVEAMVAEAKTKIVGNPFDD-VDQGPQVDKDQLNKIIDLINSGKSEGAQL 735 +V+E IYD+FVE VA AK+++VGNPFD +QGPQVD+ Q KI+ IN+GK EGA+L Sbjct: 325 TFVQEDIYDEFVERSVARAKSRVVGNPFDSKTEQGPQVDETQFKKILGYINTGKQEGAKL 384 Query: 736 CTGGKQLGDKGYFVEPTIFSKVNDNMRIAKEEIFGPVMQIMKFKNIDEVIERANNNEYGL 915 GG D+GYF++PT+F V D M IAKEEIFGPVMQI+KFK I+EV+ RANN+ YGL Sbjct: 385 LCGGGIAADRGYFIQPTVFGDVQDGMTIAKEEIFGPVMQILKFKTIEEVVGRANNSTYGL 444 Query: 916 AASVFTKDIEKSMHIMQSLRAGTVWINCYDAFDAAAPFGGYKASGVGRELGEYGLNNYSE 1095 AA+VFTKD++K+ ++ Q+L+AGTVW+NCYD F A +PFGGYK SG GRELGEYGL Y+E Sbjct: 445 AAAVFTKDLDKANYLSQALQAGTVWVNCYDVFGAQSPFGGYKMSGSGRELGEYGLQAYTE 504 Query: 1096 VKMVTIKVPSKSS 1134 VK VT+KVP K+S Sbjct: 505 VKTVTVKVPQKNS 517
>sp|P81178|ALDH2_MESAU Aldehyde dehydrogenase, mitochondrial (ALDH class 2) (ALDH1) (ALDH-E2) Length = 500 Score = 545 bits (1405), Expect = e-155 Identities = 255/373 (68%), Positives = 305/373 (81%), Gaps = 1/373 (0%) Frame = +1 Query: 19 CYEYYAGWADKNHGKVIPMRGNYLGFTRHEAVGICGQIIPWNFPLLMQAWKLGPALCMGN 198 C YYAGWADK HGK IP+ G++ +TRHE VG+CGQIIPWNFPLLMQAWKLGPAL GN Sbjct: 128 CLRYYAGWADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQAWKLGPALATGN 187 Query: 199 TVVMKTAEQTPLTANYVASLIAEAGFPPGVVNIVPGMGPVAGAALVSHPNVDKIAFTGST 378 VVMK AEQTPLTA YVA+LI EAGFPPGVVNIVPG GP AGAA+ SH +VDK+AFTGST Sbjct: 188 VVVMKVAEQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASHEDVDKVAFTGST 247 Query: 379 EVGRIISSECNKNNIKRLTLELGGKSPNIVLADADFDHAVKSSHFALFFNMGQCCCAGSR 558 EVG +I +N+KR+TLELGGKSPNI+++DAD D AV+ +HFALFFN GQCCCAGSR Sbjct: 248 EVGHLIQVAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALFFNQGQCCCAGSR 307 Query: 559 IYVEEKIYDKFVEAMVAEAKTKIVGNPFDD-VDQGPQVDKDQLNKIIDLINSGKSEGAQL 735 +V+E +YD+FVE VA AK+++VGNPFD +QGPQVD+ Q KI+ I SG+ EGA+L Sbjct: 308 TFVQEDVYDEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKILGYIKSGQQEGAKL 367 Query: 736 CTGGKQLGDKGYFVEPTIFSKVNDNMRIAKEEIFGPVMQIMKFKNIDEVIERANNNEYGL 915 GG D+GYF++PT+F V D M IAKEEIFGPVMQI+KFK I+EV+ RANN++YGL Sbjct: 368 LCGGGAAADRGYFIQPTVFGDVKDGMTIAKEEIFGPVMQILKFKTIEEVVGRANNSKYGL 427 Query: 916 AASVFTKDIEKSMHIMQSLRAGTVWINCYDAFDAAAPFGGYKASGVGRELGEYGLNNYSE 1095 AA+VFTKD++K+ ++ Q+L+AGTVWINCYD F A +PFGGYK SG GRELGEYGL Y+E Sbjct: 428 AAAVFTKDLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGSGRELGEYGLQAYTE 487 Query: 1096 VKMVTIKVPSKSS 1134 VK VTIKVP K+S Sbjct: 488 VKTVTIKVPQKNS 500
>sp|P11884|ALDH2_RAT Aldehyde dehydrogenase, mitochondrial precursor (ALDH class 2) (ALDH1) (ALDH-E2) Length = 519 Score = 545 bits (1404), Expect = e-154 Identities = 254/373 (68%), Positives = 305/373 (81%), Gaps = 1/373 (0%) Frame = +1 Query: 19 CYEYYAGWADKNHGKVIPMRGNYLGFTRHEAVGICGQIIPWNFPLLMQAWKLGPALCMGN 198 C YYAGWADK HGK IP+ G++ +TRHE VG+CGQIIPWNFPLLMQAWKLGPAL GN Sbjct: 147 CLRYYAGWADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQAWKLGPALATGN 206 Query: 199 TVVMKTAEQTPLTANYVASLIAEAGFPPGVVNIVPGMGPVAGAALVSHPNVDKIAFTGST 378 VVMK AEQTPLTA YVA+LI EAGFPPGVVNIVPG GP AGAA+ SH +VDK+AFTGST Sbjct: 207 VVVMKVAEQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASHEDVDKVAFTGST 266 Query: 379 EVGRIISSECNKNNIKRLTLELGGKSPNIVLADADFDHAVKSSHFALFFNMGQCCCAGSR 558 EVG +I +N+KR+TLELGGKSPNI+++DAD D AV+ +HFALFFN GQCCCAGSR Sbjct: 267 EVGHLIQVAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALFFNQGQCCCAGSR 326 Query: 559 IYVEEKIYDKFVEAMVAEAKTKIVGNPFDD-VDQGPQVDKDQLNKIIDLINSGKSEGAQL 735 +V+E +YD+FVE VA AK+++VGNPFD +QGPQVD+ Q KI+ I SG+ EGA+L Sbjct: 327 TFVQEDVYDEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKILGYIKSGQQEGAKL 386 Query: 736 CTGGKQLGDKGYFVEPTIFSKVNDNMRIAKEEIFGPVMQIMKFKNIDEVIERANNNEYGL 915 GG D+GYF++PT+F V D M IAKEEIFGPVMQI+KFK I+EV+ RANN++YGL Sbjct: 387 LCGGGAAADRGYFIQPTVFGDVKDGMTIAKEEIFGPVMQILKFKTIEEVVGRANNSKYGL 446 Query: 916 AASVFTKDIEKSMHIMQSLRAGTVWINCYDAFDAAAPFGGYKASGVGRELGEYGLNNYSE 1095 AA+VFTKD++K+ ++ Q+L+AGTVWINCYD F A +PFGGYK SG GRELGEYGL Y+E Sbjct: 447 AAAVFTKDLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGSGRELGEYGLQAYTE 506 Query: 1096 VKMVTIKVPSKSS 1134 VK VT+KVP K+S Sbjct: 507 VKTVTVKVPQKNS 519
>sp|P47738|ALDH2_MOUSE Aldehyde dehydrogenase, mitochondrial precursor (ALDH class 2) (AHD-M1) (ALDHI) (ALDH-E2) Length = 519 Score = 543 bits (1399), Expect = e-154 Identities = 253/373 (67%), Positives = 304/373 (81%), Gaps = 1/373 (0%) Frame = +1 Query: 19 CYEYYAGWADKNHGKVIPMRGNYLGFTRHEAVGICGQIIPWNFPLLMQAWKLGPALCMGN 198 C YYAGWADK HGK IP+ G++ +TRHE VG+CGQIIPWNFPLLMQAWKLGPAL GN Sbjct: 147 CLRYYAGWADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQAWKLGPALATGN 206 Query: 199 TVVMKTAEQTPLTANYVASLIAEAGFPPGVVNIVPGMGPVAGAALVSHPNVDKIAFTGST 378 VVMK AEQTPLTA YVA+LI EAGFPPGVVNIVPG GP AGAA+ SH VDK+AFTGST Sbjct: 207 VVVMKVAEQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASHEGVDKVAFTGST 266 Query: 379 EVGRIISSECNKNNIKRLTLELGGKSPNIVLADADFDHAVKSSHFALFFNMGQCCCAGSR 558 EVG +I +N+KR+TLELGGKSPNI+++DAD D AV+ +HFALFFN GQCCCAGSR Sbjct: 267 EVGHLIQVAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALFFNQGQCCCAGSR 326 Query: 559 IYVEEKIYDKFVEAMVAEAKTKIVGNPFDD-VDQGPQVDKDQLNKIIDLINSGKSEGAQL 735 +V+E +YD+FVE VA AK+++VGNPFD +QGPQVD+ Q KI+ I SG+ EGA+L Sbjct: 327 TFVQENVYDEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKILGYIKSGQQEGAKL 386 Query: 736 CTGGKQLGDKGYFVEPTIFSKVNDNMRIAKEEIFGPVMQIMKFKNIDEVIERANNNEYGL 915 GG D+GYF++PT+F V D M IAKEEIFGPVMQI+KFK I+EV+ RAN+++YGL Sbjct: 387 LCGGGAAADRGYFIQPTVFGDVKDGMTIAKEEIFGPVMQILKFKTIEEVVGRANDSKYGL 446 Query: 916 AASVFTKDIEKSMHIMQSLRAGTVWINCYDAFDAAAPFGGYKASGVGRELGEYGLNNYSE 1095 AA+VFTKD++K+ ++ Q+L+AGTVWINCYD F A +PFGGYK SG GRELGEYGL Y+E Sbjct: 447 AAAVFTKDLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGSGRELGEYGLQAYTE 506 Query: 1096 VKMVTIKVPSKSS 1134 VK VT+KVP K+S Sbjct: 507 VKTVTVKVPQKNS 519
>sp|P20000|ALDH2_BOVIN Aldehyde dehydrogenase, mitochondrial precursor (ALDH class 2) (ALDHI) (ALDH-E2) Length = 520 Score = 540 bits (1391), Expect = e-153 Identities = 248/373 (66%), Positives = 305/373 (81%), Gaps = 1/373 (0%) Frame = +1 Query: 19 CYEYYAGWADKNHGKVIPMRGNYLGFTRHEAVGICGQIIPWNFPLLMQAWKLGPALCMGN 198 C YYAGWADK HGK IP+ G+Y +TRHE VG+CGQIIPWNFPLLMQAWKLGPAL GN Sbjct: 148 CLRYYAGWADKYHGKTIPIDGDYFSYTRHEPVGVCGQIIPWNFPLLMQAWKLGPALATGN 207 Query: 199 TVVMKTAEQTPLTANYVASLIAEAGFPPGVVNIVPGMGPVAGAALVSHPNVDKIAFTGST 378 VVMK AEQTPLTA YVA+LI EAGFPPGVVN++PG GP AGAA+ SH +VDK+AFTGST Sbjct: 208 VVVMKVAEQTPLTALYVANLIKEAGFPPGVVNVIPGFGPTAGAAIASHEDVDKVAFTGST 267 Query: 379 EVGRIISSECNKNNIKRLTLELGGKSPNIVLADADFDHAVKSSHFALFFNMGQCCCAGSR 558 EVG +I K+N+KR+TLE+GGKSPNI+++DAD D AV+ +HFALFFN GQCCCAGSR Sbjct: 268 EVGHLIQVAAGKSNLKRVTLEIGGKSPNIIMSDADMDWAVEQAHFALFFNQGQCCCAGSR 327 Query: 559 IYVEEKIYDKFVEAMVAEAKTKIVGNPFDD-VDQGPQVDKDQLNKIIDLINSGKSEGAQL 735 +V+E IY +FVE VA AK+++VGNPFD +QGPQVD+ Q K++ I SGK EG +L Sbjct: 328 TFVQEDIYAEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKVLGYIKSGKEEGLKL 387 Query: 736 CTGGKQLGDKGYFVEPTIFSKVNDNMRIAKEEIFGPVMQIMKFKNIDEVIERANNNEYGL 915 GG D+GYF++PT+F + D M IAKEEIFGPVMQI+KFK+++EV+ RANN++YGL Sbjct: 388 LCGGGAAADRGYFIQPTVFGDLQDGMTIAKEEIFGPVMQILKFKSMEEVVGRANNSKYGL 447 Query: 916 AASVFTKDIEKSMHIMQSLRAGTVWINCYDAFDAAAPFGGYKASGVGRELGEYGLNNYSE 1095 AA+VFTKD++K+ ++ Q+L+AGTVW+NCYD F A +PFGGYK SG GRELGEYGL Y+E Sbjct: 448 AAAVFTKDLDKANYLSQALQAGTVWVNCYDVFGAQSPFGGYKLSGSGRELGEYGLQAYTE 507 Query: 1096 VKMVTIKVPSKSS 1134 VK VT++VP K+S Sbjct: 508 VKTVTVRVPQKNS 520
>sp|P12762|ALDH2_HORSE Aldehyde dehydrogenase, mitochondrial (ALDH class 2) (ALDHI) (ALDH-E2) Length = 500 Score = 532 bits (1371), Expect = e-151 Identities = 250/373 (67%), Positives = 303/373 (81%), Gaps = 1/373 (0%) Frame = +1 Query: 19 CYEYYAGWADKNHGKVIPMRGNYLGFTRHEAVGICGQIIPWNFPLLMQAWKLGPALCMGN 198 C YYAGWADK HGK IP+ G++ +TRHE VG+CGQIIPWNFPLLMQA KLGPAL GN Sbjct: 128 CLRYYAGWADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQAAKLGPALATGN 187 Query: 199 TVVMKTAEQTPLTANYVASLIAEAGFPPGVVNIVPGMGPVAGAALVSHPNVDKIAFTGST 378 VVMK AEQTPLTA YVA+L EAGFPPGVVN+VPG GP AGAA+ SH +VDK+AFTGST Sbjct: 188 VVVMKVAEQTPLTALYVANLTKEAGFPPGVVNVVPGFGPTAGAAIASHEDVDKVAFTGST 247 Query: 379 EVGRIISSECNKNNIKRLTLELGGKSPNIVLADADFDHAVKSSHFALFFNMGQCCCAGSR 558 EVG +I ++N+K++TLELGGKSPNI+++DAD D AV+ +HFALFFN GQCC AGSR Sbjct: 248 EVGHLIQVAAGRSNLKKVTLELGGKSPNIIVSDADMDWAVEQAHFALFFNQGQCCGAGSR 307 Query: 559 IYVEEKIYDKFVEAMVAEAKTKIVGNPFD-DVDQGPQVDKDQLNKIIDLINSGKSEGAQL 735 +V+E +Y +FVE VA AK+++VGNPFD +QGPQVD+ Q NK++ I SGK EGA+L Sbjct: 308 TFVQEDVYAEFVERSVARAKSRVVGNPFDSQTEQGPQVDETQFNKVLGYIKSGKEEGAKL 367 Query: 736 CTGGKQLGDKGYFVEPTIFSKVNDNMRIAKEEIFGPVMQIMKFKNIDEVIERANNNEYGL 915 GG D+GYF++PT+F V D M IAKEEIFGPVMQI+KFK I+EV+ RANN++YGL Sbjct: 368 LCGGGAAADRGYFIQPTVFGDVQDGMTIAKEEIFGPVMQILKFKTIEEVVGRANNSKYGL 427 Query: 916 AASVFTKDIEKSMHIMQSLRAGTVWINCYDAFDAAAPFGGYKASGVGRELGEYGLNNYSE 1095 AA+VFTKD++K+ ++ Q+L+AGTVWINCYD F A +PFGGYK SG GRELGEYGL Y+E Sbjct: 428 AAAVFTKDLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGNGRELGEYGLQAYTE 487 Query: 1096 VKMVTIKVPSKSS 1134 VK VTIKVP K+S Sbjct: 488 VKTVTIKVPQKNS 500
>sp|O94788|AL1A2_HUMAN Retinal dehydrogenase 2 (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2) Length = 499 Score = 523 bits (1346), Expect = e-148 Identities = 241/372 (64%), Positives = 304/372 (81%), Gaps = 1/372 (0%) Frame = +1 Query: 22 YEYYAGWADKNHGKVIPMRGNYLGFTRHEAVGICGQIIPWNFPLLMQAWKLGPALCMGNT 201 + YYAGWADK HG IP+ G+Y FTRHE +G+CGQIIPWNFPLLM AWK+ PALC GNT Sbjct: 128 FRYYAGWADKIHGMTIPVDGDYFTFTRHEPIGVCGQIIPWNFPLLMFAWKIAPALCCGNT 187 Query: 202 VVMKTAEQTPLTANYVASLIAEAGFPPGVVNIVPGMGPVAGAALVSHPNVDKIAFTGSTE 381 VV+K AEQTPL+A Y+ +LI EAGFPPGV+NI+PG GP AGAA+ SH +DKIAFTGSTE Sbjct: 188 VVIKPAEQTPLSALYMGALIKEAGFPPGVINILPGYGPTAGAAIASHIGIDKIAFTGSTE 247 Query: 382 VGRIISSECNKNNIKRLTLELGGKSPNIVLADADFDHAVKSSHFALFFNMGQCCCAGSRI 561 VG++I ++N+KR+TLELGGKSPNI+ ADAD D+AV+ +H +FFN GQCC AGSRI Sbjct: 248 VGKLIQEAAGRSNLKRVTLELGGKSPNIIFADADLDYAVEQAHQGVFFNQGQCCTAGSRI 307 Query: 562 YVEEKIYDKFVEAMVAEAKTKIVGNPFD-DVDQGPQVDKDQLNKIIDLINSGKSEGAQLC 738 +VEE IY++FV V AK +IVG+PFD +QGPQ+DK Q NKI++LI SG +EGA+L Sbjct: 308 FVEESIYEEFVRRSVERAKRRIVGSPFDPTTEQGPQIDKKQYNKILELIQSGVAEGAKLE 367 Query: 739 TGGKQLGDKGYFVEPTIFSKVNDNMRIAKEEIFGPVMQIMKFKNIDEVIERANNNEYGLA 918 GGK LG KG+F+EPT+FS V D+MRIAKEEIFGPV +I++FK +DEVIERANN+++GL Sbjct: 368 CGGKGLGRKGFFIEPTVFSNVTDDMRIAKEEIFGPVQEILRFKTMDEVIERANNSDFGLV 427 Query: 919 ASVFTKDIEKSMHIMQSLRAGTVWINCYDAFDAAAPFGGYKASGVGRELGEYGLNNYSEV 1098 A+VFT DI K++ + +++AGTVWINCY+A +A +PFGG+K SG GRE+GE+GL YSEV Sbjct: 428 AAVFTNDINKALTVSSAMQAGTVWINCYNALNAQSPFGGFKMSGNGREMGEFGLREYSEV 487 Query: 1099 KMVTIKVPSKSS 1134 K VT+K+P K+S Sbjct: 488 KTVTVKIPQKNS 499
>sp|O93344|AL1A2_CHICK Retinal dehydrogenase 2 (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2) Length = 499 Score = 522 bits (1345), Expect = e-148 Identities = 242/370 (65%), Positives = 302/370 (81%), Gaps = 1/370 (0%) Frame = +1 Query: 28 YYAGWADKNHGKVIPMRGNYLGFTRHEAVGICGQIIPWNFPLLMQAWKLGPALCMGNTVV 207 YYAGWADK HG IP+ G+Y FTRHE +G+CGQIIPWNFPLLM AWK+ PALC GNTVV Sbjct: 130 YYAGWADKIHGMTIPVDGDYFTFTRHEPIGVCGQIIPWNFPLLMFAWKIAPALCCGNTVV 189 Query: 208 MKTAEQTPLTANYVASLIAEAGFPPGVVNIVPGMGPVAGAALVSHPNVDKIAFTGSTEVG 387 +K AEQTPL+A Y+ +LI EAGFPPGVVNI+PG GP+ GAA+ SH +DKIAFTGSTEVG Sbjct: 190 IKPAEQTPLSALYMGALIKEAGFPPGVVNILPGFGPIVGAAIASHVGIDKIAFTGSTEVG 249 Query: 388 RIISSECNKNNIKRLTLELGGKSPNIVLADADFDHAVKSSHFALFFNMGQCCCAGSRIYV 567 ++I ++N+KR+TLELGGKSPNI+ ADAD D+AV+ +H +FFN GQCC AGSRIYV Sbjct: 250 KLIQEAAGRSNLKRVTLELGGKSPNIIFADADLDYAVEQAHQGVFFNQGQCCTAGSRIYV 309 Query: 568 EEKIYDKFVEAMVAEAKTKIVGNPFD-DVDQGPQVDKDQLNKIIDLINSGKSEGAQLCTG 744 EE IY++FV V AK ++VG+PFD +QGPQ+DK Q NKI++LI SG +EGA+L G Sbjct: 310 EESIYEEFVRRSVERAKRRVVGSPFDPTTEQGPQIDKKQYNKILELIQSGITEGAKLECG 369 Query: 745 GKQLGDKGYFVEPTIFSKVNDNMRIAKEEIFGPVMQIMKFKNIDEVIERANNNEYGLAAS 924 GK LG KG+F+EPT+FS V D+MRIAKEEIFGPV +I++FK +DEVIERANN+++GL A+ Sbjct: 370 GKGLGRKGFFIEPTVFSNVTDDMRIAKEEIFGPVQEILRFKTVDEVIERANNSDFGLVAA 429 Query: 925 VFTKDIEKSMHIMQSLRAGTVWINCYDAFDAAAPFGGYKASGVGRELGEYGLNNYSEVKM 1104 VFT DI K++ + +++AGTVWINCY+A +A +PFGG+K SG GRE+GE GL YSEVK Sbjct: 430 VFTNDINKALTVSSAMQAGTVWINCYNALNAQSPFGGFKMSGNGREMGESGLREYSEVKT 489 Query: 1105 VTIKVPSKSS 1134 VTIK+P K+S Sbjct: 490 VTIKIPQKNS 499
>sp|Q63639|AL1A2_RAT Retinal dehydrogenase 2 (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2) Length = 499 Score = 521 bits (1341), Expect = e-147 Identities = 241/370 (65%), Positives = 303/370 (81%), Gaps = 1/370 (0%) Frame = +1 Query: 28 YYAGWADKNHGKVIPMRGNYLGFTRHEAVGICGQIIPWNFPLLMQAWKLGPALCMGNTVV 207 YYAGWADK HG IP+ G+Y FTRHE +G+CGQIIPWNFPLLM WK+ PALC GNTVV Sbjct: 130 YYAGWADKIHGMTIPVDGDYFTFTRHEPIGVCGQIIPWNFPLLMFTWKIAPALCCGNTVV 189 Query: 208 MKTAEQTPLTANYVASLIAEAGFPPGVVNIVPGMGPVAGAALVSHPNVDKIAFTGSTEVG 387 +K AEQTPL+A Y+ +LI EAGFPPGVVNI+PG GP AGAA+ SH +DKIAFTGSTEVG Sbjct: 190 IKPAEQTPLSALYMGALIKEAGFPPGVVNILPGYGPTAGAAIASHIGIDKIAFTGSTEVG 249 Query: 388 RIISSECNKNNIKRLTLELGGKSPNIVLADADFDHAVKSSHFALFFNMGQCCCAGSRIYV 567 ++I ++N+KR+TLELGGKSPNI+ ADAD D+AV+ +H +FFN GQCC AGSRI+V Sbjct: 250 KLIQEAAGRSNLKRVTLELGGKSPNIIFADADLDYAVEQAHQGVFFNQGQCCTAGSRIFV 309 Query: 568 EEKIYDKFVEAMVAEAKTKIVGNPFD-DVDQGPQVDKDQLNKIIDLINSGKSEGAQLCTG 744 EE IY++FV+ V AK +IVG+PFD +QGPQ+DK Q NKI++LI SG +EGA+L G Sbjct: 310 EESIYEEFVKRSVERAKRRIVGSPFDPTTEQGPQIDKKQYNKILELIQSGVAEGAKLECG 369 Query: 745 GKQLGDKGYFVEPTIFSKVNDNMRIAKEEIFGPVMQIMKFKNIDEVIERANNNEYGLAAS 924 GK LG KG+F+EPT+FS V D+MRIAKEEIFGPV +I++FK +DEVIERANN+++GL A+ Sbjct: 370 GKGLGRKGFFIEPTVFSNVTDDMRIAKEEIFGPVQEILRFKTMDEVIERANNSDFGLVAA 429 Query: 925 VFTKDIEKSMHIMQSLRAGTVWINCYDAFDAAAPFGGYKASGVGRELGEYGLNNYSEVKM 1104 VFT DI K++ + +++AGTVWINCY+A +A +PFGG+K SG GRE+GE+GL YSEVK Sbjct: 430 VFTNDINKALMVSSAMQAGTVWINCYNALNAQSPFGGFKMSGNGREMGEFGLREYSEVKT 489 Query: 1105 VTIKVPSKSS 1134 VT+K+P K+S Sbjct: 490 VTVKIPQKNS 499
>sp|Q62148|AL1A2_MOUSE Retinal dehydrogenase 2 (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2) Length = 499 Score = 520 bits (1340), Expect = e-147 Identities = 240/370 (64%), Positives = 303/370 (81%), Gaps = 1/370 (0%) Frame = +1 Query: 28 YYAGWADKNHGKVIPMRGNYLGFTRHEAVGICGQIIPWNFPLLMQAWKLGPALCMGNTVV 207 YYAGWADK HG IP+ G+Y FTRHE +G+CGQIIPWNFPLLM WK+ PALC GNTVV Sbjct: 130 YYAGWADKIHGMTIPVDGDYFTFTRHEPIGVCGQIIPWNFPLLMFTWKIAPALCCGNTVV 189 Query: 208 MKTAEQTPLTANYVASLIAEAGFPPGVVNIVPGMGPVAGAALVSHPNVDKIAFTGSTEVG 387 +K AEQTPL+A Y+ +LI EAGFPPGVVNI+PG GP AGAA+ SH +DKIAFTGSTEVG Sbjct: 190 IKPAEQTPLSALYMGALIKEAGFPPGVVNILPGYGPTAGAAIASHIGIDKIAFTGSTEVG 249 Query: 388 RIISSECNKNNIKRLTLELGGKSPNIVLADADFDHAVKSSHFALFFNMGQCCCAGSRIYV 567 ++I ++N+KR+TLELGGKSPNI+ ADAD D+AV+ +H +FFN GQCC AGSRI+V Sbjct: 250 KLIQEAAGRSNLKRVTLELGGKSPNIIFADADLDYAVEQAHQGVFFNQGQCCTAGSRIFV 309 Query: 568 EEKIYDKFVEAMVAEAKTKIVGNPFD-DVDQGPQVDKDQLNKIIDLINSGKSEGAQLCTG 744 EE IY++FV+ V AK +IVG+PFD +QGPQ+DK Q NK+++LI SG +EGA+L G Sbjct: 310 EESIYEEFVKRSVERAKRRIVGSPFDPTTEQGPQIDKKQYNKVLELIQSGVAEGAKLECG 369 Query: 745 GKQLGDKGYFVEPTIFSKVNDNMRIAKEEIFGPVMQIMKFKNIDEVIERANNNEYGLAAS 924 GK LG KG+F+EPT+FS V D+MRIAKEEIFGPV +I++FK +DEVIERANN+++GL A+ Sbjct: 370 GKGLGRKGFFIEPTVFSNVTDDMRIAKEEIFGPVQEILRFKTMDEVIERANNSDFGLVAA 429 Query: 925 VFTKDIEKSMHIMQSLRAGTVWINCYDAFDAAAPFGGYKASGVGRELGEYGLNNYSEVKM 1104 VFT DI K++ + +++AGTVWINCY+A +A +PFGG+K SG GRE+GE+GL YSEVK Sbjct: 430 VFTNDINKALMVSSAMQAGTVWINCYNALNAQSPFGGFKMSGNGREMGEFGLREYSEVKT 489 Query: 1105 VTIKVPSKSS 1134 VT+K+P K+S Sbjct: 490 VTVKIPQKNS 499
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 143,644,069
Number of Sequences: 369166
Number of extensions: 2932843
Number of successful extensions: 8446
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7625
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7938
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 15134460800
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail