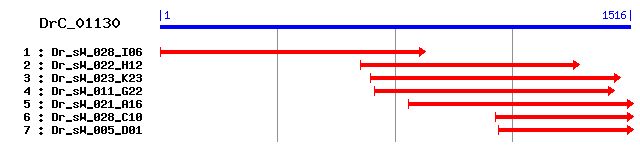
DrC_01130
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01130 (1516 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9ULA0|DNPEP_HUMAN Aspartyl aminopeptidase 491 e-138 sp|Q9Z2W0|DNPEP_MOUSE Aspartyl aminopeptidase 474 e-133 sp|Q19087|DNPEP_CAEEL Putative aspartyl aminopeptidase 417 e-116 sp|O36014|DNPEP_SCHPO Putative aspartyl aminopeptidase 397 e-110 sp|P38821|DNPEP_YEAST Putative aspartyl aminopeptidase 379 e-104 sp|Q9HYZ3|APEB_PSEAE Probable M18-family aminopeptidase 2 331 4e-90 sp|Q88M44|APEB_PSEPK Probable M18-family aminopeptidase 2 328 3e-89 sp|Q87YC5|APEB_PSESM Probable M18-family aminopeptidase 2 328 3e-89 sp|Q97LF4|APEB_CLOAB Probable M18-family aminopeptidase 2 300 1e-80 sp|Q82F74|APEB_STRAW Probable M18-family aminopeptidase 2 244 6e-64
>sp|Q9ULA0|DNPEP_HUMAN Aspartyl aminopeptidase Length = 475 Score = 491 bits (1265), Expect = e-138 Identities = 237/461 (51%), Positives = 333/461 (72%), Gaps = 5/461 (1%) Frame = +2 Query: 20 AKNLIKFIDKSPTPYHAVASVKDMLVNYGFKEIHEQDEWNLKPNDLVFVTKNHSTIIALA 199 AK L+KF+++SP+P+HAVA ++ L+ GF E+ E ++WN+KP F+T+N STIIA A Sbjct: 18 AKELLKFVNRSPSPFHAVAECRNRLLQAGFSELKETEKWNIKPESKYFMTRNSSTIIAFA 77 Query: 200 IGGKYKPGNGFSMIGAHTDSPCLRLKPCSEQLSNTYMKLGVETYGGGLWYTWFDRDLTVA 379 +GG+Y PGNGFS+IGAHTDSPCLR+K S + + ++GVETYGGG+W TWFDRDLT+A Sbjct: 78 VGGQYVPGNGFSLIGAHTDSPCLRVKRRSRRSQVGFQQVGVETYGGGIWSTWFDRDLTLA 137 Query: 380 GRCVVR--ESDKYRHLLVNLHRPIARIPTLAVHLDRSVNDSFAPNKELHLNPVLCTTIYE 553 GR +V+ S + LV++ RPI RIP LA+HL R++N++F PN E+HL P+L T I E Sbjct: 138 GRVIVKCPTSGRLEQQLVHVERPILRIPHLAIHLQRNINENFGPNTEMHLVPILATAIQE 197 Query: 554 QINKINSEQKDSPTSSLNHPTVLLEAIAQELKVKVEDIVDVELCLADCQPSTIGGIHQEF 733 ++ K E H +VL+ + L + +DIV++ELCLAD QP+ +GG + EF Sbjct: 198 ELEKGTPEPGPLNAVDERHHSVLMSLLCAHLGLSPKDIVEMELCLADTQPAVLGGAYDEF 257 Query: 734 VFAPRLDNLFNCYTSVQALVESLE---SVSDDTNIRAVCLFDNEEIGSQSAQGAQSAFTE 904 +FAPRLDNL +C+ ++QAL++S S++ + ++R V L+DNEE+GS+SAQGAQS TE Sbjct: 258 IFAPRLDNLHSCFCALQALIDSCAGPGSLATEPHVRMVTLYDNEEVGSESAQGAQSLLTE 317 Query: 905 LILRRVVNTICQNCGVASFSHYERAIANSLLISADQAHGVHPNYEEKHERQHKPLPHQGI 1084 L+LRR+ + CQ+ + +E AI S +ISAD AH VHPNY +KHE H+PL H+G Sbjct: 318 LVLRRI-SASCQHP-----TAFEEAIPKSFMISADMAHAVHPNYLDKHEENHRPLFHKGP 371 Query: 1085 VLKINSNQRYASNAMTTSILREVANKSEVPLQNFVVRQDMACGSTIGPIMSANMGLATVD 1264 V+K+NS QRYASNA++ +++REVANK +VPLQ+ +VR D CG+TIGPI+++ +GL +D Sbjct: 372 VIKVNSKQRYASNAVSEALIREVANKVKVPLQDLMVRNDTPCGTTIGPILASRLGLRVLD 431 Query: 1265 VGGPQLSMHSCREMTDVSSVEQAVHLFKGFFNLYPDLSRRV 1387 +G PQL+MHS REM + V Q + LFKGFF L+P LS + Sbjct: 432 LGSPQLAMHSIREMACTTGVLQTLTLFKGFFELFPSLSHNL 472
>sp|Q9Z2W0|DNPEP_MOUSE Aspartyl aminopeptidase Length = 473 Score = 474 bits (1221), Expect = e-133 Identities = 226/461 (49%), Positives = 326/461 (70%), Gaps = 5/461 (1%) Frame = +2 Query: 20 AKNLIKFIDKSPTPYHAVASVKDMLVNYGFKEIHEQDEWNLKPNDLVFVTKNHSTIIALA 199 A+ L+KF+++SP+P+H VA + L+ GF+E+ E + W++ P + F+T+N S+IIA A Sbjct: 16 ARELLKFVNRSPSPFHVVAECRSRLLQAGFRELKETEGWDIVPENNYFLTRNSSSIIAFA 75 Query: 200 IGGKYKPGNGFSMIGAHTDSPCLRLKPCSEQLSNTYMKLGVETYGGGLWYTWFDRDLTVA 379 +GG+Y PGNGFS+IGAHTDSPCLR+K S + Y ++GVETYGGG+W TWFDRDLT+A Sbjct: 76 VGGQYVPGNGFSLIGAHTDSPCLRVKRKSRRSQVGYHQVGVETYGGGIWSTWFDRDLTLA 135 Query: 380 GRCVVR--ESDKYRHLLVNLHRPIARIPTLAVHLDRSVNDSFAPNKELHLNPVLCTTIYE 553 GR +++ S + LV++ RPI RIP LA+HL R++N++F PN E+HL P+L T + E Sbjct: 136 GRVIIKCPTSGRLEQRLVHIERPILRIPHLAIHLQRNINENFGPNTEIHLVPILATAVQE 195 Query: 554 QINKINSEQKDSPTSSLNHPTVLLEAIAQELKVKVEDIVDVELCLADCQPSTIGGIHQEF 733 ++ K E + H +VL+ + L + + I+++ELCLAD QP+ +GG ++EF Sbjct: 196 ELEKGTPEPGPLGATDERHHSVLMSLLCTHLGLSPDSIMEMELCLADTQPAVLGGAYEEF 255 Query: 734 VFAPRLDNLFNCYTSVQALVESLES---VSDDTNIRAVCLFDNEEIGSQSAQGAQSAFTE 904 +FAPRLDNL +C+ ++QAL++S S ++ D ++R V L+DNEE+GS+SAQGAQS TE Sbjct: 256 IFAPRLDNLHSCFCALQALIDSCASPASLARDPHVRMVTLYDNEEVGSESAQGAQSLLTE 315 Query: 905 LILRRVVNTICQNCGVASFSHYERAIANSLLISADQAHGVHPNYEEKHERQHKPLPHQGI 1084 LILRR+ + + +E AI S +ISAD AH VHPNY +KHE H+P H+G Sbjct: 316 LILRRI------SASPQRLTAFEEAIPKSFMISADMAHAVHPNYSDKHEENHRPSFHKGP 369 Query: 1085 VLKINSNQRYASNAMTTSILREVANKSEVPLQNFVVRQDMACGSTIGPIMSANMGLATVD 1264 V+K+NS QRYASNA++ S++REVA + VPLQ+ +VR D CG+TIGPI+++ +GL +D Sbjct: 370 VIKVNSKQRYASNAVSESMIREVAGQVGVPLQDLMVRNDSPCGTTIGPILASRLGLRVLD 429 Query: 1265 VGGPQLSMHSCREMTDVSSVEQAVHLFKGFFNLYPDLSRRV 1387 +G PQL+MHS RE + V Q + LFKGFF L+P +SR + Sbjct: 430 LGSPQLAMHSIRETACTTGVLQTLTLFKGFFELFPSVSRNL 470
>sp|Q19087|DNPEP_CAEEL Putative aspartyl aminopeptidase Length = 470 Score = 417 bits (1072), Expect = e-116 Identities = 212/452 (46%), Positives = 289/452 (63%), Gaps = 6/452 (1%) Frame = +2 Query: 20 AKNLIKFIDKSPTPYHAVASVKDMLVNYGFKEIHEQDEWNLKPNDLVFVTKNHSTIIALA 199 A+ I +++K+ TP+HA VKD L+ GF E+ E W+++P FVTKN S I+A A Sbjct: 16 AQEFINYLNKAVTPFHATQEVKDRLLQAGFTELPESGHWDIQPTSKYFVTKNRSAILAFA 75 Query: 200 IGGKYKPGNGFSMIGAHTDSPCLRLKPCSEQLSNTYMKLGVETYGGGLWYTWFDRDLTVA 379 +GG YKPG+GFS++ HTDSPCLR+KP S Q S+ ++++GV TYGGG+W TWFDRDL+VA Sbjct: 76 VGGSYKPGSGFSIVVGHTDSPCLRVKPISHQKSDKFLQVGVSTYGGGIWRTWFDRDLSVA 135 Query: 380 GRCVVRESDKYRHLLVNLHRPIARIPTLAVHLDRSVNDSFAPNKELHLNPVLCTTIYEQI 559 G +V+ +K +H L+++ +P+ IP LA+HL+ +F PN E L P+L T I Sbjct: 136 GLVIVKNGEKLQHKLIDVKKPVLFIPNLAIHLETD-RTTFKPNTETELRPILETFAAAGI 194 Query: 560 NKINSEQK----DSPTSSLNHPTVLLEAIAQELKVKVEDIVDVELCLADCQPSTIGGIHQ 727 N + D + NH L IA+E + EDIVD++L L D + I G+ Sbjct: 195 NAPQKPESTGFADPRNITNNHHPQFLGLIAKEAGCQPEDIVDLDLYLYDTNKAAIVGMED 254 Query: 728 EFVFAPRLDNLFNCYTSVQALVESL--ESVSDDTNIRAVCLFDNEEIGSQSAQGAQSAFT 901 EF+ RLDN YT++ L+ESL ES +D IR FDNEE+GS SA GA S+FT Sbjct: 255 EFISGARLDNQVGTYTAISGLLESLTGESFKNDPQIRIAACFDNEEVGSDSAMGASSSFT 314 Query: 902 ELILRRVVNTICQNCGVASFSHYERAIANSLLISADQAHGVHPNYEEKHERQHKPLPHQG 1081 E +LRR+ S + +E AI S+LISADQAH HPNY KHE H+P H G Sbjct: 315 EFVLRRL-------SAGGSTTAFEEAIGKSMLISADQAHATHPNYSAKHEENHRPAFHGG 367 Query: 1082 IVLKINSNQRYASNAMTTSILREVANKSEVPLQNFVVRQDMACGSTIGPIMSANMGLATV 1261 +V+K+N NQRYA+ + T + L++VA +++VPLQ VVR D CGST+GPI++ +GL TV Sbjct: 368 VVVKVNVNQRYATTSTTHAALKQVAFEAQVPLQVVVVRNDSPCGSTVGPILATKLGLQTV 427 Query: 1262 DVGGPQLSMHSCREMTDVSSVEQAVHLFKGFF 1357 DVG PQL+MHS RE D SS+ QA L+ F+ Sbjct: 428 DVGCPQLAMHSIREFADTSSIYQATTLYSTFY 459
>sp|O36014|DNPEP_SCHPO Putative aspartyl aminopeptidase Length = 467 Score = 397 bits (1021), Expect = e-110 Identities = 214/475 (45%), Positives = 311/475 (65%), Gaps = 12/475 (2%) Frame = +2 Query: 5 TSLNYAKNLIKFIDKSPTPYHAVASVKDMLVNYGFKEIHEQDEWNLK--PNDLVFVTKNH 178 T+ + A + + F++ SPTPYHAV ++ + +++GF+ + E+ +W K P + FVT+N Sbjct: 4 TAKSCALDFLDFVNASPTPYHAVQNLAEHYMSHGFQYLSEKSDWQSKIEPGNSYFVTRNK 63 Query: 179 STIIALAIGGKYKPGNGFSMIGAHTDSPCLRLKPCSEQLSNTYMKLGVETYGGGLWYTWF 358 S+IIA +IG K+KPGNGFS+I HTDSP LRLKP S++ + Y+++GVE YGGG+W+TWF Sbjct: 64 SSIIAFSIGKKWKPGNGFSIIATHTDSPTLRLKPKSQKSAYGYLQVGVEKYGGGIWHTWF 123 Query: 359 DRDLTVAGRCVVRESDKYRHLLVNLH--RPIARIPTLAVHLDRSVNDSFAPNKELHLNPV 532 DRDL++AGR +V E D R + N+H RP+ RIPTLA+HLD S N SF+ N E P+ Sbjct: 124 DRDLSLAGRVMVEEEDG-RVIQYNVHIDRPLLRIPTLAIHLDPSANSSFSFNMETEFVPL 182 Query: 533 LCTTIYEQINKINSEQKDSPTSSLNHPTVLLEAIAQELKVKVED------IVDVELCLAD 694 + N++ E+ +HP VLL +A E+ +E IVD EL L D Sbjct: 183 IGLE-----NELAKEETSDNGDKYHHP-VLLSLLANEISKSLETTIDPSKIVDFELILGD 236 Query: 695 CQPSTIGGIHQEFVFAPRLDNLFNCYTSVQALVESLE--SVSDDTNIRAVCLFDNEEIGS 868 + + +GGIH+EFVF+PRLDNL + + QAL +SLE S+ +++ +R V FD+EEIGS Sbjct: 237 AEKARLGGIHEEFVFSPRLDNLGMTFCASQALTKSLENNSLDNESCVRVVPSFDHEEIGS 296 Query: 869 QSAQGAQSAFTELILRRVVNTICQNCGVASFSHYERAIANSLLISADQAHGVHPNYEEKH 1048 SAQGA+S F +L+R IC+ G S S + ++ S L+SAD AH +HPNY ++ Sbjct: 297 VSAQGAESTFLPAVLQR----ICE-LGKES-SLFSISMVKSFLVSADMAHAMHPNYSSRY 350 Query: 1049 ERQHKPLPHQGIVLKINSNQRYASNAMTTSILREVANKSEVPLQNFVVRQDMACGSTIGP 1228 E + P ++G V+K+N+NQRY +N+ +L++VA ++VP+Q+FVVR D CGSTIGP Sbjct: 351 ENSNTPFLNKGTVIKVNANQRYTTNSAGIVLLKKVAQLADVPIQSFVVRNDSPCGSTIGP 410 Query: 1229 IMSANMGLATVDVGGPQLSMHSCREMTDVSSVEQAVHLFKGFFNLYPDLSRRVTL 1393 ++A G+ T+D+G P LSMHSCREM E AV LF FF + +L ++ + Sbjct: 411 KLAAMTGMRTLDLGNPMLSMHSCREMCGSKDFEYAVVLFSSFFQNFANLEEKIII 465
>sp|P38821|DNPEP_YEAST Putative aspartyl aminopeptidase Length = 490 Score = 379 bits (972), Expect = e-104 Identities = 211/478 (44%), Positives = 300/478 (62%), Gaps = 18/478 (3%) Frame = +2 Query: 14 NYAKNLIKFIDKSPTPYHAVASVKDMLVNYGFKEIHEQDEW--NLKPNDLVFVTKNHSTI 187 +Y K + F++ S +PYH V ++K LV+ GFKE+ E+D W ++ FVT+N S+I Sbjct: 17 DYPKEFVSFLNSSHSPYHTVHNIKKHLVSNGFKELSERDSWAGHVAQKGKYFVTRNGSSI 76 Query: 188 IALAIGGKYKPGNGFSMIGAHTDSPCLRLKPCSEQLSNTYMKLGVETYGGGLWYTWFDRD 367 IA A+GGK++PGN ++ GAHTDSP LR+KP S+++S Y+++GVETYGG +W++WFD+D Sbjct: 77 IAFAVGGKWEPGNPIAITGAHTDSPALRIKPISKRVSEKYLQVGVETYGGAIWHSWFDKD 136 Query: 368 LTVAGRCVVRESDKYRHL--LVNLHRPIARIPTLAVHLDRSVNDSFAPNKELHLNPV--- 532 L VAGR V+++ + + LV+L+RP+ +IPTLA+HLDR VN F N+E L P+ Sbjct: 137 LGVAGRVFVKDAKTGKSIARLVDLNRPLLKIPTLAIHLDRDVNQKFEFNRETQLLPIGGL 196 Query: 533 ----LCTTIYEQINKINSEQKDSPTSSLNHPTVLLEAIAQELKVK-VEDIVDVELCLADC 697 ++IN N E T H LL IA+EL + +EDI D EL L D Sbjct: 197 QEDKTEAKTEKEIN--NGEFTSIKTIVQRHHAELLGLIAKELAIDTIEDIEDFELILYDH 254 Query: 698 QPSTIGGIHQEFVFAPRLDNLFNCYTSVQALVESLESVSD-DTNIRAVCLFDNEEIGSQS 874 ST+GG + EFVF+ RLDNL +C+TS+ L + ++ D ++ IR + FD+EEIGS S Sbjct: 255 NASTLGGFNDEFVFSGRLDNLTSCFTSMHGLTLAADTEIDRESGIRLMACFDHEEIGSSS 314 Query: 875 AQGAQSAFTELILRRVVNTICQNCGVASFSH-YERAI----ANSLLISADQAHGVHPNYE 1039 AQGA S F IL R+ +I + G + AI A S +S+D AH VHPNY Sbjct: 315 AQGADSNFLPNILERL--SILKGDGSDQTKPLFHSAILETSAKSFFLSSDVAHAVHPNYA 372 Query: 1040 EKHERQHKPLPHQGIVLKINSNQRYASNAMTTSILREVANKSEVPLQNFVVRQDMACGST 1219 K+E QHKPL G V+KIN+NQRY +N+ +++ +A ++VPLQ FVV D CGST Sbjct: 373 NKYESQHKPLLGGGPVIKINANQRYMTNSPGLVLVKRLAEAAKVPLQLFVVANDSPCGST 432 Query: 1220 IGPIMSANMGLATVDVGGPQLSMHSCREMTDVSSVEQAVHLFKGFFNLYPDLSRRVTL 1393 IGPI+++ G+ T+D+G P LSMHS RE + +E + LFK FF Y + + + Sbjct: 433 IGPILASKTGIRTLDLGNPVLSMHSIRETGGSADLEFQIKLFKEFFERYTSIESEIVV 490
>sp|Q9HYZ3|APEB_PSEAE Probable M18-family aminopeptidase 2 Length = 429 Score = 331 bits (848), Expect = 4e-90 Identities = 175/445 (39%), Positives = 261/445 (58%) Frame = +2 Query: 23 KNLIKFIDKSPTPYHAVASVKDMLVNYGFKEIHEQDEWNLKPNDLVFVTKNHSTIIALAI 202 + LI F+ SPTP+HA AS+ L G++ + E+D W+ + +VT+N S++IA+ + Sbjct: 7 QGLIDFLKASPTPFHATASLARRLEAAGYRRLDERDAWHTETGGRYYVTRNDSSLIAIRL 66 Query: 203 GGKYKPGNGFSMIGAHTDSPCLRLKPCSEQLSNTYMKLGVETYGGGLWYTWFDRDLTVAG 382 G + +GF ++GAHTDSPCLR+KP E N +++LGVE YGG L+ WFDRDL++AG Sbjct: 67 GRRSPLESGFRLVGAHTDSPCLRVKPNPEIARNGFLQLGVEVYGGALFAPWFDRDLSLAG 126 Query: 383 RCVVRESDKYRHLLVNLHRPIARIPTLAVHLDRSVNDSFAPNKELHLNPVLCTTIYEQIN 562 R R + K LV+ + IA IP LA+HL+R+ N+ + N + L P++ Sbjct: 127 RVTFRANGKLESRLVDFRKAIAVIPNLAIHLNRAANEGWPINAQNELPPIIAQL------ 180 Query: 563 KINSEQKDSPTSSLNHPTVLLEAIAQELKVKVEDIVDVELCLADCQPSTIGGIHQEFVFA 742 +P + + +L E + +E + + ++D EL D Q + + G++ EF+ Sbjct: 181 --------APGEAADFRLLLDEQLLREHGITADVVLDYELSFYDTQSAAVVGLNDEFIAG 232 Query: 743 PRLDNLFNCYTSVQALVESLESVSDDTNIRAVCLFDNEEIGSQSAQGAQSAFTELILRRV 922 RLDNL +C+ ++AL+ + D N VC D+EE+GS S GA F E +LRR+ Sbjct: 233 ARLDNLLSCHAGLEALL----NAEGDENCILVCT-DHEEVGSCSHCGADGPFLEQVLRRL 287 Query: 923 VNTICQNCGVASFSHYERAIANSLLISADQAHGVHPNYEEKHERQHKPLPHQGIVLKINS 1102 + + RAI SLL+SAD AHGVHPNY ++H+ H P + G V+KINS Sbjct: 288 ---------LPEGDAFSRAIQRSLLVSADNAHGVHPNYADRHDANHGPALNGGPVIKINS 338 Query: 1103 NQRYASNAMTTSILREVANKSEVPLQNFVVRQDMACGSTIGPIMSANMGLATVDVGGPQL 1282 NQRYA+N+ T R + SEVP+Q+FV R DM CGSTIGPI ++ +G+ TVD+G P Sbjct: 339 NQRYATNSETAGFFRHLCQDSEVPVQSFVTRSDMGCGSTIGPITASQVGVRTVDIGLPTF 398 Query: 1283 SMHSCREMTDVSSVEQAVHLFKGFF 1357 +MHS RE+ + V + F+ Sbjct: 399 AMHSIRELAGSHDLAHLVKVLGAFY 423
>sp|Q88M44|APEB_PSEPK Probable M18-family aminopeptidase 2 Length = 429 Score = 328 bits (841), Expect = 3e-89 Identities = 177/443 (39%), Positives = 262/443 (59%) Frame = +2 Query: 29 LIKFIDKSPTPYHAVASVKDMLVNYGFKEIHEQDEWNLKPNDLVFVTKNHSTIIALAIGG 208 LI+F+ SPTP+HA AS+ L G+K + E+D W+ + +VT+N S+IIA+ +G Sbjct: 9 LIEFLKASPTPFHATASLVQRLEAAGYKRLDERDSWDTETGGRYYVTRNDSSIIAIKLGK 68 Query: 209 KYKPGNGFSMIGAHTDSPCLRLKPCSEQLSNTYMKLGVETYGGGLWYTWFDRDLTVAGRC 388 + NG M+GAHTDSPCLR+KP E +++LGVE YGG L WFDRDL++AGR Sbjct: 69 QSPLLNGIRMVGAHTDSPCLRVKPQPELQRQGFLQLGVEVYGGALLAPWFDRDLSLAGRV 128 Query: 389 VVRESDKYRHLLVNLHRPIARIPTLAVHLDRSVNDSFAPNKELHLNPVLCTTIYEQINKI 568 R K L++ P+A IP LA+HL+R+ N+ +A N + L P+L ++ Sbjct: 129 TYRRDGKVESQLIDFKLPVAIIPNLAIHLNRTANEGWAINPQNELPPILAQVAGDE---- 184 Query: 569 NSEQKDSPTSSLNHPTVLLEAIAQELKVKVEDIVDVELCLADCQPSTIGGIHQEFVFAPR 748 ++ +L E +A+E ++ + ++D EL D Q + + G++ +F+ R Sbjct: 185 ----------RIDFRALLTEQLAREHELIADVVLDYELSFYDTQDAALIGLNGDFIAGAR 234 Query: 749 LDNLFNCYTSVQALVESLESVSDDTNIRAVCLFDNEEIGSQSAQGAQSAFTELILRRVVN 928 LDNL +CY +QAL L + SD+T + VC D+EE+GS SA GA E L+R+ Sbjct: 235 LDNLLSCYAGLQAL---LAADSDETCV-LVC-NDHEEVGSCSACGADGPMLEQTLQRL-- 287 Query: 929 TICQNCGVASFSHYERAIANSLLISADQAHGVHPNYEEKHERQHKPLPHQGIVLKINSNQ 1108 + Y R + SL++SAD AHGVHPNY +KH+ H P + G V+K+N+NQ Sbjct: 288 -------LPDGDTYVRTVQRSLMVSADNAHGVHPNYADKHDGNHGPKLNAGPVIKVNNNQ 340 Query: 1109 RYASNAMTTSILREVANKSEVPLQNFVVRQDMACGSTIGPIMSANMGLATVDVGGPQLSM 1288 RYA+N+ T R + EVP+Q+FVVR DM CGSTIGPI ++++G+ TVD+G P +M Sbjct: 341 RYATNSETAGFFRHLCMAEEVPVQSFVVRSDMGCGSTIGPITASHLGVRTVDIGLPTFAM 400 Query: 1289 HSCREMTDVSSVEQAVHLFKGFF 1357 HS RE+ + V + F+ Sbjct: 401 HSIRELCGSHDLAHLVKVLTAFY 423
>sp|Q87YC5|APEB_PSESM Probable M18-family aminopeptidase 2 Length = 429 Score = 328 bits (840), Expect = 3e-89 Identities = 177/445 (39%), Positives = 259/445 (58%) Frame = +2 Query: 23 KNLIKFIDKSPTPYHAVASVKDMLVNYGFKEIHEQDEWNLKPNDLVFVTKNHSTIIALAI 202 K LI F+ SPTP+HA A++ GF+ + E+D W ++ +VT+N S+I+A+ + Sbjct: 7 KGLIDFLKASPTPFHATATLVQHFEAAGFQRLDERDTWAIETGGRYYVTRNDSSIVAIRM 66 Query: 203 GGKYKPGNGFSMIGAHTDSPCLRLKPCSEQLSNTYMKLGVETYGGGLWYTWFDRDLTVAG 382 G + G M+GAHTDSPCLR+KP E + +LGVE YGG L WFDRDL++AG Sbjct: 67 GRQSPLTGGIRMVGAHTDSPCLRVKPQPELQRQGFWQLGVEVYGGALLAPWFDRDLSLAG 126 Query: 383 RCVVRESDKYRHLLVNLHRPIARIPTLAVHLDRSVNDSFAPNKELHLNPVLCTTIYEQIN 562 R R K L++ PIA IP LA+HL+R+ N+ + N + L P+L ++ Sbjct: 127 RVTFRRDGKVESQLIDFKLPIAVIPNLAIHLNRTANEGWTINPQNELPPILAQVAGDE-- 184 Query: 563 KINSEQKDSPTSSLNHPTVLLEAIAQELKVKVEDIVDVELCLADCQPSTIGGIHQEFVFA 742 + +L + +A+E + + ++D EL D Q + + G++ +F+ Sbjct: 185 ------------RADFRALLTDQLAREHGLNADVVLDYELSFYDTQSAAVVGLNGDFLAG 232 Query: 743 PRLDNLFNCYTSVQALVESLESVSDDTNIRAVCLFDNEEIGSQSAQGAQSAFTELILRRV 922 RLDNL +C+ +QAL L + SD+T + VC D+EE+GS SA GA A E I++R+ Sbjct: 233 ARLDNLLSCFAGMQAL---LNTESDETAL-LVCT-DHEEVGSSSACGADGAMLEQIVQRL 287 Query: 923 VNTICQNCGVASFSHYERAIANSLLISADQAHGVHPNYEEKHERQHKPLPHQGIVLKINS 1102 + S Y R I SLLISAD AHG+HPNY +KH+ H P + G V+K+NS Sbjct: 288 ---------LPSSEDYVRTIQKSLLISADNAHGIHPNYADKHDANHGPKLNAGPVIKVNS 338 Query: 1103 NQRYASNAMTTSILREVANKSEVPLQNFVVRQDMACGSTIGPIMSANMGLATVDVGGPQL 1282 NQRYA+N+ T R + EVP+Q+FVVR DM CGSTIGPI ++++G+ TVD+G P Sbjct: 339 NQRYATNSETAGFFRHLCMAEEVPVQSFVVRSDMGCGSTIGPITASHLGIRTVDIGLPTF 398 Query: 1283 SMHSCREMTDVSSVEQAVHLFKGFF 1357 +MHS RE+ + V + F+ Sbjct: 399 AMHSIRELAGSHDLAHLVKVLGAFY 423
>sp|Q97LF4|APEB_CLOAB Probable M18-family aminopeptidase 2 Length = 433 Score = 300 bits (767), Expect = 1e-80 Identities = 178/454 (39%), Positives = 257/454 (56%), Gaps = 3/454 (0%) Frame = +2 Query: 11 LNYAKNLIKFIDKSPTPYHAVASVKDMLVNYGFKEIHEQDEWNLKPNDLVFVTKNHSTII 190 L +AK LI FI SP+P+H+V ++K+ L+ GF EI E+++W L N FV +N S +I Sbjct: 5 LEFAKKLIDFIYDSPSPFHSVDNIKNTLIENGFAEIKEENKWELNKNGKYFVKRNDSALI 64 Query: 191 ALAIGGKYKPGNGFSMIGAHTDSPCLRLKPCSEQLS-NTYMKLGVETYGGGLWYTWFDRD 367 A +G + GF +IG HTDSP R+KP E +S N+Y+KL E YGG + TWFDR Sbjct: 65 AFTVGSGFVAKKGFKIIGGHTDSPTFRIKPNPEMVSENSYIKLNTEVYGGPILSTWFDRP 124 Query: 368 LTVAGRCVVRESDKY--RHLLVNLHRPIARIPTLAVHLDRSVNDSFAPNKELHLNPVLCT 541 L++AGR VR L+N+ RPI IP LA+H++R VN F N ++ P++ Sbjct: 125 LSIAGRVTVRGKSALFPETKLLNIKRPILVIPNLAIHMNRDVNSGFKINPQVDTLPII-G 183 Query: 542 TIYEQINKINSEQKDSPTSSLNHPTVLLEAIAQELKVKVEDIVDVELCLADCQPSTIGGI 721 I ++ K N L++ IA EL +E+I+D +L L + I GI Sbjct: 184 IINDKFEKEN---------------YLMKIIASELGEDIENIIDFDLFLYEYDKGCIMGI 228 Query: 722 HQEFVFAPRLDNLFNCYTSVQALVESLESVSDDTNIRAVCLFDNEEIGSQSAQGAQSAFT 901 + EF+ + RLD++ + + ALV + S+ TN+ A FDNEEIGS + QGA S F Sbjct: 229 NNEFISSSRLDDMEMVHAGLNALVNA--KCSEATNVLAC--FDNEEIGSATKQGADSQFL 284 Query: 902 ELILRRVVNTICQNCGVASFSHYERAIANSLLISADQAHGVHPNYEEKHERQHKPLPHQG 1081 IL R+V + + RA+ NS +IS+D AH VHPN EK + +P ++G Sbjct: 285 SDILERIVLSFG-----GDREDFFRALHNSFMISSDSAHAVHPNKGEKADPITRPHINEG 339 Query: 1082 IVLKINSNQRYASNAMTTSILREVANKSEVPLQNFVVRQDMACGSTIGPIMSANMGLATV 1261 V+KI++ Q+Y S++ + ++ EV S VP Q FV R D GSTIGPI + + + TV Sbjct: 340 PVIKISAAQKYTSDSNSIAVYEEVCRLSGVPYQKFVNRSDERGGSTIGPITATHTAIRTV 399 Query: 1262 DVGGPQLSMHSCREMTDVSSVEQAVHLFKGFFNL 1363 D+G P L+MHS RE+ F+ F+NL Sbjct: 400 DIGTPLLAMHSIRELCGTLDHMYVEKSFEEFYNL 433
>sp|Q82F74|APEB_STRAW Probable M18-family aminopeptidase 2 Length = 432 Score = 244 bits (622), Expect = 6e-64 Identities = 147/431 (34%), Positives = 218/431 (50%), Gaps = 1/431 (0%) Frame = +2 Query: 17 YAKNLIKFIDKSPTPYHAVASVKDMLVNYGFKEIHEQDEWNLKPNDLVFVTKNHSTIIAL 196 + +L+ F+ SP+PYHAV + + L GF+++ E D W+ V +IA Sbjct: 11 HTDDLMSFLAASPSPYHAVTNAAERLEKAGFRQVAETDAWDSTSGGKYVV--RGGALIAW 68 Query: 197 AIGGKYKPGNGFSMIGAHTDSPCLRLKPCSEQLSNTYMKLGVETYGGGLWYTWFDRDLTV 376 + P F ++GAHTDSP LR+KP + ++ + ++ VE YGG L +W DRDL + Sbjct: 69 YVPEGADPHTPFRIVGAHTDSPNLRVKPRPDSGAHGWRQIAVEIYGGPLLNSWLDRDLGL 128 Query: 377 AGRCVVRESDKYRHLLVNLHRPIARIPTLAVHLDRSVN-DSFAPNKELHLNPVLCTTIYE 553 AGR +R+ LVN+ RP+ R+P LA+HLDRSV+ D +K+ HL P+ Sbjct: 129 AGRLSLRDGSTR---LVNIDRPLLRVPQLAIHLDRSVSTDGLKLDKQRHLQPIW------ 179 Query: 554 QINKINSEQKDSPTSSLNHPTVLLEAIAQELKVKVEDIVDVELCLADCQPSTIGGIHQEF 733 + + +D L+ + EL + ++ +L + + G E Sbjct: 180 ---GLGGDVRDGD---------LIAFLEAELGLSAGEVTGWDLMTHSVEAPSYLGRDNEL 227 Query: 734 VFAPRLDNLFNCYTSVQALVESLESVSDDTNIRAVCLFDNEEIGSQSAQGAQSAFTELIL 913 + PR+DNL + + AL + + I + FD+EE GSQS GA +L Sbjct: 228 LAGPRMDNLLSVHAGTAALTAVAAADASLAYIPVLAAFDHEENGSQSDTGADGPLLGSVL 287 Query: 914 RRVVNTICQNCGVASFSHYERAIANSLLISADQAHGVHPNYEEKHERQHKPLPHQGIVLK 1093 R V S+ RA A S+ +S+D H VHPNY E+H+ H P G +LK Sbjct: 288 ERSVFARS-----GSYEDRARAFAGSVCLSSDTGHAVHPNYAERHDPTHHPRAGGGPILK 342 Query: 1094 INSNQRYASNAMTTSILREVANKSEVPLQNFVVRQDMACGSTIGPIMSANMGLATVDVGG 1273 +N N RYA++ +I K+ VP Q FV M CG+TIGPI +A G+ TVD+G Sbjct: 343 VNVNNRYATDGSGRAIFAAACEKANVPFQTFVSNNSMPCGTTIGPITAARHGIRTVDIGA 402 Query: 1274 PQLSMHSCREM 1306 LSMHS RE+ Sbjct: 403 AILSMHSAREL 413
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 176,927,149
Number of Sequences: 369166
Number of extensions: 3850206
Number of successful extensions: 9746
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9071
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9673
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 18325966995
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail