DrC_01067
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01067 (1827 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q5R8Y6|TM9S2_PONPY Transmembrane 9 superfamily protein m... 684 0.0 sp|Q99805|TM9S2_HUMAN Transmembrane 9 superfamily protein m... 683 0.0 sp|P58021|TM9S2_MOUSE Transmembrane 9 superfamily protein m... 682 0.0 sp|Q66HG5|TM9S2_RAT Transmembrane 9 superfamily protein mem... 681 0.0 sp|Q92544|TM9S4_HUMAN Transmembrane 9 superfamily protein m... 470 e-132 sp|P32802|EMP70_YEAST Endosomal P24A protein precursor (70 ... 375 e-103 sp|Q9DBU0|TM9S1_MOUSE Transmembrane 9 superfamily protein m... 280 1e-74 sp|O15321|TM9S1_HUMAN Transmembrane 9 superfamily protein m... 279 2e-74 sp|Q5R8F1|TM9S1_PONPY Transmembrane 9 superfamily protein m... 277 6e-74 sp|Q9HD45|TM9S3_HUMAN Transmembrane 9 superfamily protein m... 268 3e-71
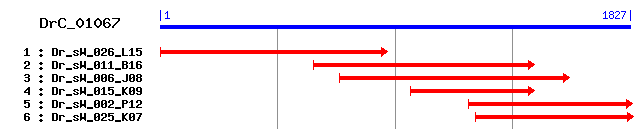
>sp|Q5R8Y6|TM9S2_PONPY Transmembrane 9 superfamily protein member 2 precursor Length = 663 Score = 684 bits (1766), Expect = 0.0 Identities = 330/574 (57%), Positives = 417/574 (72%), Gaps = 6/574 (1%) Frame = +3 Query: 12 NLGQILFGERIQNSPYKIEFGKDLSYKEVCQKKYSKNDAS-QSKLKFLMSGIMLNYRHHW 188 NLGQ+LFGERI+ SPYK F K+ + K VC K Y A + KL+FL ++LNY+HHW Sbjct: 95 NLGQVLFGERIEPSPYKFTFNKEETCKLVCTKTYHTEKAEDKQKLEFLKKSMLLNYQHHW 154 Query: 189 IVDDLPVVSCYK-SGSNEVCYNGFPIGCYQSDKAKSV-MCTTDASQNIPNTYFLFNHISL 362 IVD++PV CY C GFPIGCY +DK + C ++ + +T+++FNH+ Sbjct: 155 IVDNMPVTWCYDVEDGQRFCNPGFPIGCYITDKGHAKDACVINSEFHERDTFYIFNHVD- 213 Query: 363 TIGIYTDEINEKLSKGRIVYVRVNTASKAFVKEGATDKELANAPPMIL--KGNEDTVIRY 536 I IY + R+V ++ S K DK + PPM + K + + I Y Sbjct: 214 -IKIYYHVVETGSMGARLVAAKLEPKS---FKHTHIDKPDCSGPPMDISNKASGEIKIAY 269 Query: 537 TYSVKFEPTNEIKWSSRWDHVLKTMPQSNIQWFSILNSVIIVLFLSGMLAMILLKTLHRD 716 TYSV FE +I+W+SRWD++L++MP ++IQWFSI+NS++IVLFLSGM+AMI+L+TLH+D Sbjct: 270 TYSVSFEEDKKIRWASRWDYILESMPHTHIQWFSIMNSLVIVLFLSGMVAMIMLRTLHKD 329 Query: 717 IDRYNKLD-TEEAQEEFGWKLVHGDVFRPPRMGMTLCVFVGSGVQLILMTIVTLFISCFG 893 I RYN++D TE+AQEEFGWKLVHGD+FRPPR GM L VF+GSG Q+++MT VTLF +C G Sbjct: 330 IARYNQMDSTEDAQEEFGWKLVHGDIFRPPRKGMLLSVFLGSGTQILIMTFVTLFFACLG 389 Query: 894 FLSPANRGAFATCGIVFYVLFGAFAGFYSARLYKMMSGEKWKSNVLLTAFLCPGSVFGIF 1073 FLSPANRGA TC +V +VL G AG+ +AR YK GEKWK+NVLLT+FLCPG VF F Sbjct: 390 FLSPANRGALMTCAVVLWVLLGTPAGYVAARFYKSFGGEKWKTNVLLTSFLCPGIVFADF 449 Query: 1074 FVLNIVLWASGSSAAVPFTTILGLLAMWFLISVPLCFVGAYFGFKKQPISQPVRTNQIPR 1253 F++N++LW GSSAA+PF T++ +LA+WF ISVPL F+GAYFGFKK I PVRTNQIPR Sbjct: 450 FIMNLILWGEGSSAAIPFGTLVAILALWFCISVPLTFIGAYFGFKKNAIEHPVRTNQIPR 509 Query: 1254 QIPYQYLYTRSIPSILMGGVLPFGCIFIQLFFILNSIWSTYVYYMFGXXXXXXXXXXXTC 1433 QIP Q YT+ +P I+MGG+LPFGCIFIQLFFILNSIWS +YYMFG TC Sbjct: 510 QIPEQSFYTKPLPGIIMGGILPFGCIFIQLFFILNSIWSHQMYYMFGFLFLVFIILVITC 569 Query: 1434 SETTILLCYFHLCAEDYRWWWRSFLTSGCTALYLFIYSIYFFHAKLDIKGSTSTFLYFGY 1613 SE TILLCYFHLCAEDY W WRSFLTSG TA+Y IY++++F +KL I G+ ST LYFGY Sbjct: 570 SEATILLCYFHLCAEDYHWQWRSFLTSGFTAVYFLIYAVHYFFSKLQITGTASTILYFGY 629 Query: 1614 TAIIIWLSFLVTGTVGFVSSFWFVRKIYSVVKVD 1715 T I++ + FL TGT+GF + FWFV KIYSVVKVD Sbjct: 630 TMIMVLIFFLFTGTIGFFACFWFVTKIYSVVKVD 663
>sp|Q99805|TM9S2_HUMAN Transmembrane 9 superfamily protein member 2 precursor (p76) Length = 663 Score = 683 bits (1763), Expect = 0.0 Identities = 330/574 (57%), Positives = 416/574 (72%), Gaps = 6/574 (1%) Frame = +3 Query: 12 NLGQILFGERIQNSPYKIEFGKDLSYKEVCQKKYSKNDAS-QSKLKFLMSGIMLNYRHHW 188 NLGQ+LFGERI+ SPYK F K + K VC K Y A + KL+FL ++LNY+HHW Sbjct: 95 NLGQVLFGERIEPSPYKFTFNKKETCKLVCTKTYHTEKAEDKQKLEFLKKSMLLNYQHHW 154 Query: 189 IVDDLPVVSCYK-SGSNEVCYNGFPIGCYQSDKAKSV-MCTTDASQNIPNTYFLFNHISL 362 IVD++PV CY C GFPIGCY +DK + C + + +T+++FNH+ Sbjct: 155 IVDNMPVTWCYDVEDGQRFCNPGFPIGCYITDKGHAKDACVISSDFHERDTFYIFNHVD- 213 Query: 363 TIGIYTDEINEKLSKGRIVYVRVNTASKAFVKEGATDKELANAPPMIL--KGNEDTVIRY 536 I IY + R+V ++ S K DK + PPM + K + + I Y Sbjct: 214 -IKIYYHVVETGSMGARLVAAKLEPKS---FKHTHIDKPDCSGPPMDISNKASGEIKIAY 269 Query: 537 TYSVKFEPTNEIKWSSRWDHVLKTMPQSNIQWFSILNSVIIVLFLSGMLAMILLKTLHRD 716 TYSV FE ++I+W+SRWD++L++MP ++IQWFSI+NS++IVLFLSGM+AMI+L+TLH+D Sbjct: 270 TYSVSFEEDDKIRWASRWDYILESMPHTHIQWFSIMNSLVIVLFLSGMVAMIMLRTLHKD 329 Query: 717 IDRYNKLD-TEEAQEEFGWKLVHGDVFRPPRMGMTLCVFVGSGVQLILMTIVTLFISCFG 893 I RYN++D TE+AQEEFGWKLVHGD+FRPPR GM L VF+GSG Q+++MT VTLF +C G Sbjct: 330 IARYNQMDSTEDAQEEFGWKLVHGDIFRPPRKGMLLSVFLGSGTQILIMTFVTLFFACLG 389 Query: 894 FLSPANRGAFATCGIVFYVLFGAFAGFYSARLYKMMSGEKWKSNVLLTAFLCPGSVFGIF 1073 FLSPANRGA TC +V +VL G AG+ +AR YK GEKWK+NVLLT+FLCPG VF F Sbjct: 390 FLSPANRGALMTCAVVLWVLLGTPAGYVAARFYKSFGGEKWKTNVLLTSFLCPGIVFADF 449 Query: 1074 FVLNIVLWASGSSAAVPFTTILGLLAMWFLISVPLCFVGAYFGFKKQPISQPVRTNQIPR 1253 F++N++LW GSSAA+PF T++ +LA+WF ISVPL F+GAYFGFKK I PVRTNQIPR Sbjct: 450 FIMNLILWGEGSSAAIPFGTLVAILALWFCISVPLTFIGAYFGFKKNAIEHPVRTNQIPR 509 Query: 1254 QIPYQYLYTRSIPSILMGGVLPFGCIFIQLFFILNSIWSTYVYYMFGXXXXXXXXXXXTC 1433 QIP Q YT+ +P I+MGG+LPFGCIFIQLFFILNSIWS +YYMFG TC Sbjct: 510 QIPEQSFYTKPLPGIIMGGILPFGCIFIQLFFILNSIWSHQMYYMFGFLFLVFIILVITC 569 Query: 1434 SETTILLCYFHLCAEDYRWWWRSFLTSGCTALYLFIYSIYFFHAKLDIKGSTSTFLYFGY 1613 SE TILLCYFHLCAEDY W WRSFLTSG TA+Y IY++++F +KL I G+ ST LYFGY Sbjct: 570 SEATILLCYFHLCAEDYHWQWRSFLTSGFTAVYFLIYAVHYFFSKLQITGTASTILYFGY 629 Query: 1614 TAIIIWLSFLVTGTVGFVSSFWFVRKIYSVVKVD 1715 T I++ + FL TGT+GF + FWFV KIYSVVKVD Sbjct: 630 TMIMVLIFFLFTGTIGFFACFWFVTKIYSVVKVD 663
>sp|P58021|TM9S2_MOUSE Transmembrane 9 superfamily protein member 2 precursor Length = 662 Score = 682 bits (1760), Expect = 0.0 Identities = 330/574 (57%), Positives = 416/574 (72%), Gaps = 6/574 (1%) Frame = +3 Query: 12 NLGQILFGERIQNSPYKIEFGKDLSYKEVCQKKYSKNDAS-QSKLKFLMSGIMLNYRHHW 188 NLGQ+LFGERI+ SPYK F K + K VC K Y+ A + KL FL ++LNY+HHW Sbjct: 94 NLGQVLFGERIEPSPYKFTFNKKETCKLVCTKTYNTEKAEDKQKLDFLKKSMLLNYQHHW 153 Query: 189 IVDDLPVVSCYK-SGSNEVCYNGFPIGCYQSDKAKSV-MCTTDASQNIPNTYFLFNHISL 362 IVD++PV CY+ S + C GFPIGCY +DK + C + + +T+++FNH+ Sbjct: 154 IVDNMPVTWCYEVEDSQKFCNPGFPIGCYITDKGHAKDACVISSEFHERDTFYIFNHVD- 212 Query: 363 TIGIYTDEINEKLSKGRIVYVRVNTASKAFVKEGATDKELANAPPMIL--KGNEDTVIRY 536 I IY + R+V ++ S K DK + P M + K + + I Y Sbjct: 213 -IKIYYHVVETGSMGARLVAAKLEPKS---FKHTHIDKPDCSGPAMDISNKASGEIKIAY 268 Query: 537 TYSVKFEPTNEIKWSSRWDHVLKTMPQSNIQWFSILNSVIIVLFLSGMLAMILLKTLHRD 716 TYS+ FE I+W+SRWD++L++MP ++IQWFSI+NS++IVLFLSGM+AMI+L+TLH+D Sbjct: 269 TYSISFEEEKNIRWASRWDYILESMPHTHIQWFSIMNSLVIVLFLSGMVAMIMLRTLHKD 328 Query: 717 IDRYNKLD-TEEAQEEFGWKLVHGDVFRPPRMGMTLCVFVGSGVQLILMTIVTLFISCFG 893 I RYN++D TE+AQEEFGWKLVHGD+FRPPR GM L VF+GSG Q+++MT VTLF +C G Sbjct: 329 IARYNQMDSTEDAQEEFGWKLVHGDIFRPPRKGMLLSVFLGSGTQILIMTFVTLFFACLG 388 Query: 894 FLSPANRGAFATCGIVFYVLFGAFAGFYSARLYKMMSGEKWKSNVLLTAFLCPGSVFGIF 1073 FLSPANRGA TC +V +VL G AG+ +AR YK GEKWK+NVLLT+FLCPG VF F Sbjct: 389 FLSPANRGALMTCAVVLWVLLGTPAGYVAARFYKSFGGEKWKTNVLLTSFLCPGIVFADF 448 Query: 1074 FVLNIVLWASGSSAAVPFTTILGLLAMWFLISVPLCFVGAYFGFKKQPISQPVRTNQIPR 1253 F++N++LW GSSAA+PF T++ +LA+WF ISVPL F+GAYFGFKK I PVRTNQIPR Sbjct: 449 FIMNLILWGEGSSAAIPFGTLVAILALWFCISVPLTFIGAYFGFKKNAIEHPVRTNQIPR 508 Query: 1254 QIPYQYLYTRSIPSILMGGVLPFGCIFIQLFFILNSIWSTYVYYMFGXXXXXXXXXXXTC 1433 QIP Q YT+ +P I+MGG+LPFGCIFIQLFFILNSIWS +YYMFG TC Sbjct: 509 QIPEQSFYTKPLPGIIMGGILPFGCIFIQLFFILNSIWSHQMYYMFGFLFLVFIILVITC 568 Query: 1434 SETTILLCYFHLCAEDYRWWWRSFLTSGCTALYLFIYSIYFFHAKLDIKGSTSTFLYFGY 1613 SE TILLCYFHLCAEDY W WRSFLTSG TA+Y IY+I++F +KL I G+ ST LYFGY Sbjct: 569 SEATILLCYFHLCAEDYHWQWRSFLTSGFTAVYFLIYAIHYFFSKLQITGTASTILYFGY 628 Query: 1614 TAIIIWLSFLVTGTVGFVSSFWFVRKIYSVVKVD 1715 T I++ + FL TGT+GF + FWFV KIYSVVKVD Sbjct: 629 TMIMVLIFFLFTGTIGFFACFWFVTKIYSVVKVD 662
>sp|Q66HG5|TM9S2_RAT Transmembrane 9 superfamily protein member 2 precursor Length = 663 Score = 681 bits (1756), Expect = 0.0 Identities = 327/574 (56%), Positives = 416/574 (72%), Gaps = 6/574 (1%) Frame = +3 Query: 12 NLGQILFGERIQNSPYKIEFGKDLSYKEVCQKKYSKNDAS-QSKLKFLMSGIMLNYRHHW 188 NLGQ+LFGERI+ SPYK F K+ + K VC K Y A + KL FL ++LNY+HHW Sbjct: 95 NLGQVLFGERIEPSPYKFTFNKEETCKLVCTKTYHTEKAEDKQKLDFLKKSMLLNYQHHW 154 Query: 189 IVDDLPVVSCYKSGSNE-VCYNGFPIGCYQSDKAKSV-MCTTDASQNIPNTYFLFNHISL 362 IVD++PV CY+ N+ C GFPIGCY +DK + C + + +T+++FNH+ + Sbjct: 155 IVDNMPVTWCYEVEDNQKFCNPGFPIGCYITDKGHAKDACVISSEFHERDTFYIFNHVDI 214 Query: 363 TIGIYTDEINEKLSKGRIVYVRVNTASKAFVKEGATDKELANAPPMIL--KGNEDTVIRY 536 I + E R+V ++ S + DK + P M + K + + I Y Sbjct: 215 KIQYHVVETGSM--GARLVAAKLEPKS---FRHTHIDKPDCSGPAMDISNKASGEIKIAY 269 Query: 537 TYSVKFEPTNEIKWSSRWDHVLKTMPQSNIQWFSILNSVIIVLFLSGMLAMILLKTLHRD 716 TYS+ FE I+W+SRWD++L++MP ++IQWFSI+NS++IVLFLSGM+AMI+L+TLH+D Sbjct: 270 TYSISFEEEKNIRWASRWDYILESMPHTHIQWFSIMNSLVIVLFLSGMVAMIMLRTLHKD 329 Query: 717 IDRYNKLD-TEEAQEEFGWKLVHGDVFRPPRMGMTLCVFVGSGVQLILMTIVTLFISCFG 893 I RYN++D TE+AQEEFGWKLVHGD+FRPPR GM L VF+GSG Q+++MT VTLF +C G Sbjct: 330 IARYNQMDSTEDAQEEFGWKLVHGDIFRPPRKGMLLSVFLGSGTQILIMTFVTLFFACLG 389 Query: 894 FLSPANRGAFATCGIVFYVLFGAFAGFYSARLYKMMSGEKWKSNVLLTAFLCPGSVFGIF 1073 FLSPANRGA TC +V +VL G AG+ +AR YK GEKWK+NVLLT+FLCPG VF F Sbjct: 390 FLSPANRGALMTCAVVLWVLLGTPAGYVAARFYKSFGGEKWKTNVLLTSFLCPGIVFADF 449 Query: 1074 FVLNIVLWASGSSAAVPFTTILGLLAMWFLISVPLCFVGAYFGFKKQPISQPVRTNQIPR 1253 F++N++LW GSSAA+PF T++ +LA+WF ISVPL F+GAYFGFKK I PVRTNQIPR Sbjct: 450 FIMNLILWGEGSSAAIPFGTLVAILALWFCISVPLTFIGAYFGFKKNAIEHPVRTNQIPR 509 Query: 1254 QIPYQYLYTRSIPSILMGGVLPFGCIFIQLFFILNSIWSTYVYYMFGXXXXXXXXXXXTC 1433 QIP Q YT+ +P I+MGG+LPFGCIFIQLFFILNSIWS +YYMFG TC Sbjct: 510 QIPEQSFYTKPLPGIIMGGILPFGCIFIQLFFILNSIWSHQMYYMFGFLFLVFIILVITC 569 Query: 1434 SETTILLCYFHLCAEDYRWWWRSFLTSGCTALYLFIYSIYFFHAKLDIKGSTSTFLYFGY 1613 SE TILLCYFHLCAEDY W WRSFLTSG TA+Y +Y+I++F +KL I G+ ST LYFGY Sbjct: 570 SEATILLCYFHLCAEDYHWQWRSFLTSGFTAVYFLVYAIHYFFSKLQITGTASTILYFGY 629 Query: 1614 TAIIIWLSFLVTGTVGFVSSFWFVRKIYSVVKVD 1715 T I++ + FL TGT+GF + FWFV KIYSVVKVD Sbjct: 630 TMIMVLIFFLFTGTIGFFACFWFVTKIYSVVKVD 663
>sp|Q92544|TM9S4_HUMAN Transmembrane 9 superfamily protein member 4 Length = 625 Score = 470 bits (1210), Expect = e-132 Identities = 245/580 (42%), Positives = 347/580 (59%), Gaps = 12/580 (2%) Frame = +3 Query: 12 NLGQILFGERIQNSPYKIEFGKDLSYKEVCQKKYSKNDASQSKLKFLMSGIMLNYRHHWI 191 NLG++L G+RI N+P+++ + + +C + + + + + I +Y H I Sbjct: 58 NLGEVLRGDRIVNTPFQVLMNSEKKCEVLCSQSNKPVTLTVEQSRLVAERITEDYYVHLI 117 Query: 192 VDDLPVVSCYKSGSNEVCYNGFPIGCYQSDKAKSVMCTTDASQNIP----NTYFLFNHIS 359 D+LPV + + SN DK K + + N +L NH+S Sbjct: 118 ADNLPVATRLELYSNRD----------SDDKKKEKDVQFEHGYRLGFTDVNKIYLHNHLS 167 Query: 360 LTIGIYTDEINEKLSKG-RIVYVRVNTAS------KAFVKEGATDKELANAPPMILKGNE 518 + + +++ E R+V V S KA K T E N+ P + + Sbjct: 168 FILYYHREDMEEDQEHTYRVVRFEVIPQSIRLEDLKADEKSSCTLPEGTNSSPQEIDPTK 227 Query: 519 DTVIRYTYSVKFEPTNEIKWSSRWDHVLKTMPQSNIQWFSILNSVIIVLFLSGMLAMILL 698 + + +TYSV +E + +IKW+SRWD L TM I WFSI+NSV++V FLSG+L+MI++ Sbjct: 228 ENQLYFTYSVHWEES-DIKWASRWDTYL-TMSDVQIHWFSIINSVVVVFFLSGILSMIII 285 Query: 699 KTLHRDIDRYNKLDT-EEAQEEFGWKLVHGDVFRPPRMGMTLCVFVGSGVQLILMTIVTL 875 +TL +DI YNK D E+ EE GWKLVHGDVFRPP+ M L +GSG+QL M ++ + Sbjct: 286 RTLRKDIANYNKEDDIEDTMEESGWKLVHGDVFRPPQYPMILSSLLGSGIQLFCMILIVI 345 Query: 876 FISCFGFLSPANRGAFATCGIVFYVLFGAFAGFYSARLYKMMSGEKWKSNVLLTAFLCPG 1055 F++ G LSP++RGA T ++ G F GF + RLY+ + G +WK TA L PG Sbjct: 346 FVAMLGMLSPSSRGALMTTACFLFMFMGVFGGFSAGRLYRTLKGHRWKKGAFCTATLYPG 405 Query: 1056 SVFGIFFVLNIVLWASGSSAAVPFTTILGLLAMWFLISVPLCFVGAYFGFKKQPISQPVR 1235 VFGI FVLN +W SS AVPF T++ LL MWF IS+PL ++G YFGF+KQP PVR Sbjct: 406 VVFGICFVLNCFIWGKHSSGAVPFPTMVALLCMWFGISLPLVYLGYYFGFRKQPYDNPVR 465 Query: 1236 TNQIPRQIPYQYLYTRSIPSILMGGVLPFGCIFIQLFFILNSIWSTYVYYMFGXXXXXXX 1415 TNQIPRQIP Q Y ILM G+LPFG +FI+LFFI ++IW YY+FG Sbjct: 466 TNQIPRQIPEQRWYMNRFVGILMAGILPFGAMFIELFFIFSAIWENQFYYLFGFLFLVFI 525 Query: 1416 XXXXTCSETTILLCYFHLCAEDYRWWWRSFLTSGCTALYLFIYSIYFFHAKLDIKGSTST 1595 +CS+ +I++ YF LCAEDYRWWWR+FL SG +A Y+ +Y+I++F KLDI + Sbjct: 526 ILVVSCSQISIVMVYFQLCAEDYRWWWRNFLVSGGSAFYVLVYAIFYFVNKLDIVEFIPS 585 Query: 1596 FLYFGYTAIIIWLSFLVTGTVGFVSSFWFVRKIYSVVKVD 1715 LYFGYTA+++ +L+TGT+GF +++ FVRKIY+ VK+D Sbjct: 586 LLYFGYTALMVLSFWLLTGTIGFYAAYMFVRKIYAAVKID 625
>sp|P32802|EMP70_YEAST Endosomal P24A protein precursor (70 kDa endomembrane protein) (Pheromone alpha-factor transporter) (Acidic 24 kDa late endocytic intermediate component) Length = 667 Score = 375 bits (962), Expect = e-103 Identities = 217/590 (36%), Positives = 325/590 (55%), Gaps = 22/590 (3%) Frame = +3 Query: 12 NLGQILFGERIQNSPYKIEFGKDLSYKEVCQKKYSKNDASQSKLKFLMSGIMLNYRHHWI 191 +LG ++FG+RI NSP+++ ++ + +C+ +DA KF+ I + +W+ Sbjct: 94 SLGSVIFGDRIYNSPFQLNMLQEKECESLCKTVIPGDDA-----KFINKLIKNGFFQNWL 148 Query: 192 VDDLPVVSCYKSGSNEVCYNG--FPIGCYQSDKAKSVMCT--------------TDASQN 323 +D LP G + + G F +G Q + + T T +N Sbjct: 149 IDGLPAAREVYDGRTKTSFYGAGFNLGFVQVTQGTDIEATPKGAETTDKDVELETRNDRN 208 Query: 324 IPNTY---FLFNHISLTIGIYTDEINEKLSKGRIVYVRVNTASKAFVKEGATDKELANAP 494 + TY + NH + I + + R+V V V S G E +P Sbjct: 209 MVKTYELPYFANHFDIMIEYH----DRGEGNYRVVGVIVEPVSIKRSSPGTC--ETTGSP 262 Query: 495 PMILKGNEDTVIRYTYSVKFEPTNEIKWSSRWDHVLKTMPQSNIQWFSILNSVIIVLFLS 674 M+ +GN++ V +TYSVKF + W++RWD L S IQWFS++N ++V+ LS Sbjct: 263 LMLDEGNDNEVY-FTYSVKFNES-ATSWATRWDKYLHVYDPS-IQWFSLINFSLVVVLLS 319 Query: 675 GMLAMILLKTLHRDIDRYNKLDTEEA-QEEFGWKLVHGDVFRPPRMGMTLCVFVGSGVQL 851 ++ LL+ L D RYN+L+ ++ QE+ GWKL HGDVFR P +TL + VGSGVQL Sbjct: 320 SVVIHSLLRALKSDFARYNELNLDDDFQEDSGWKLNHGDVFRSPSQSLTLSILVGSGVQL 379 Query: 852 ILMTIVTLFISCFGFLSPANRGAFATCGIVFYVLFGAFAGFYSARLYKMMSGEKWKSNVL 1031 LM ++F + GFLSP++RG+ AT + Y LFG + S +YK +G WK+N++ Sbjct: 380 FLMVTCSIFFAALGFLSPSSRGSLATVMFILYALFGFVGSYTSMGIYKFFNGPYWKANLI 439 Query: 1032 LTAFLCPGSVFGIFFVLNIVLWASGSSAAVPFTTILGLLAMWFLISVPLCFVGAYFGFKK 1211 LT L PG++ I LN L SS +P +T+ ++ +WFL S+PL F G+ K+ Sbjct: 440 LTPLLVPGAILLIIIALNFFLMFVHSSGVIPASTLFFMVFLWFLFSIPLSFAGSLIARKR 499 Query: 1212 QPISQ-PVRTNQIPRQIPYQYLYTRSIPSILMGGVLPFGCIFIQLFFILNSIWSTYVYYM 1388 + P +TNQI RQIP+Q Y ++IP+ L+ G+ PFG I ++L+FI S+W ++YM Sbjct: 500 CHWDEHPTKTNQIARQIPFQPWYLKTIPATLIAGIFPFGSIAVELYFIYTSLWFNKIFYM 559 Query: 1389 FGXXXXXXXXXXXTCSETTILLCYFHLCAEDYRWWWRSFLTSGC-TALYLFIYSIYFFHA 1565 FG T S TIL+ Y LC E+++W WR F+ G ALY+FI+SI F Sbjct: 560 FGFLFFSFLLLTLTSSLVTILITYHSLCLENWKWQWRGFIIGGAGCALYVFIHSILF--T 617 Query: 1566 KLDIKGSTSTFLYFGYTAIIIWLSFLVTGTVGFVSSFWFVRKIYSVVKVD 1715 K + G T+ LY GY+++I L LVTG++GF+SS FVRKIYS +KVD Sbjct: 618 KFKLGGFTTIVLYVGYSSVISLLCCLVTGSIGFISSMLFVRKIYSSIKVD 667
>sp|Q9DBU0|TM9S1_MOUSE Transmembrane 9 superfamily protein member 1 precursor Length = 606 Score = 280 bits (716), Expect = 1e-74 Identities = 180/583 (30%), Positives = 296/583 (50%), Gaps = 15/583 (2%) Frame = +3 Query: 12 NLGQILFGERIQNSPYKIEFGKDLSYKEVCQKKYSKNDASQSKLKFLMSGIMLNYRHHWI 191 +LG++L G+R+ S Y+I F +++ + +C + S Q L I Y ++ Sbjct: 78 SLGEVLDGDRMAESLYEIRFRENVEKRILCHMQLSSAQVEQ-----LRQAIEELYYFEFV 132 Query: 192 VDDLPVVSCYKSGSNEVCYNGFPIGCYQSDKAKSVMCTTDASQNIPNTYF--LFNHISLT 365 VDDLP+ GF +G + S +P+++ L+ H+ Sbjct: 133 VDDLPI-------------RGF-VGYMEE------------SGFLPHSHKIGLWTHLDFH 166 Query: 366 IGIYTDEINEKLSKGRIVYVRVNTASKAFVKEGATDKELANAPPMILKGNEDTVIRYTYS 545 + + D RI++ V+ VK + D L+ +E + +TYS Sbjct: 167 LEFHGD---------RIIFANVSVRD---VKPHSLDG---------LRSDELLGLTHTYS 205 Query: 546 VKFEPTN-EIKWSSRWDHVLKTMPQS-NIQWFSILNSVIIVLFLSGMLAMILLKTLHRDI 719 V++ T+ E + R P++ I W SI+NS+++V L G +A+IL++ L D+ Sbjct: 206 VRWSETSVEHRSDRRRGDDGGFFPRTLEIHWLSIINSMVLVFLLVGFVAVILMRVLRNDL 265 Query: 720 DRYNKLDTEEA---------QEEFGWKLVHGDVFR-PPRMGMTLCVFVGSGVQLILMTIV 869 RYN LD E + Q + GWK++H DVFR PP G+ LC +G G Q + + Sbjct: 266 ARYN-LDEETSSGGSSDDFDQGDNGWKIIHTDVFRFPPYRGL-LCAVLGVGAQFLALGTG 323 Query: 870 TLFISCFGFLSPANRGAFATCGIVFYVLFGAFAGFYSARLYKMMSGEKWKSNVLLTAFLC 1049 + ++ G + GA + I+ Y L +G+ S+ Y+ + GE+W N++LT+ L Sbjct: 324 IIVMALLGMFNVHRHGAINSAAILLYALTCCISGYVSSHFYRQIGGERWVWNIILTSSLF 383 Query: 1050 PGSVFGIFFVLNIVLWASGSSAAVPFTTILGLLAMWFLISVPLCFVGAYFGFKK-QPISQ 1226 F + V+N V WA+GS+ A+P TTIL LL +W L+ PL +G FG P Sbjct: 384 SVPFFLTWSVVNSVHWANGSTQALPATTILLLLTVWLLVGFPLTVIGGIFGKNNASPFDA 443 Query: 1227 PVRTNQIPRQIPYQYLYTRSIPSILMGGVLPFGCIFIQLFFILNSIWSTYVYYMFGXXXX 1406 P RT I R+IP Q Y ++ + +GG LPF I ++L++I ++W Y ++G Sbjct: 444 PCRTKNIAREIPPQPWYKSTVIHMTVGGFLPFSAISVELYYIFATVWGREQYTLYGILFF 503 Query: 1407 XXXXXXXTCSETTILLCYFHLCAEDYRWWWRSFLTSGCTALYLFIYSIYFFHAKLDIKGS 1586 + +I L YF L EDYRWWWRS L+ G T L++F+YS++++ + ++ G+ Sbjct: 504 VFAILLSVGACISIALTYFQLSGEDYRWWWRSVLSVGSTGLFIFLYSVFYYARRSNMSGA 563 Query: 1587 TSTFLYFGYTAIIIWLSFLVTGTVGFVSSFWFVRKIYSVVKVD 1715 T +FGY+ + ++ FL+ GT+ F SS F+R IY +K+D Sbjct: 564 VQTVEFFGYSLLTGYVFFLMLGTISFFSSLKFIRYIYVNLKMD 606
>sp|O15321|TM9S1_HUMAN Transmembrane 9 superfamily protein member 1 precursor (hMP70) Length = 606 Score = 279 bits (713), Expect = 2e-74 Identities = 180/586 (30%), Positives = 294/586 (50%), Gaps = 18/586 (3%) Frame = +3 Query: 12 NLGQILFGERIQNSPYKIEFGKDLSYKEVCQKKYSKNDASQSKLKFLMSGIMLNYRHHWI 191 +LG++L G+R+ S Y+I F +++ + +C + S Q L I Y ++ Sbjct: 78 SLGEVLDGDRMAESLYEIRFRENVEKRILCHMQLSSAQVEQ-----LRQAIEELYYFEFV 132 Query: 192 VDDLPVVSCYKSGSNEVCYNGFPIGCYQSDKAKSVMCTTDASQNIPNTYF--LFNHISLT 365 VDDLP+ GF +G + S +P+++ L+ H+ Sbjct: 133 VDDLPI-------------RGF-VGYMEE------------SGFLPHSHKIGLWTHLDFH 166 Query: 366 IGIYTDEINEKLSKGRIVYVRVNTASKAFVKEGATDKELANAPPMILKGNEDTVIRYTYS 545 + + D RI++ V+ VK + D L+ +E + +TYS Sbjct: 167 LEFHGD---------RIIFANVSVRD---VKPHSLDG---------LRPDEFLGLTHTYS 205 Query: 546 VKFEPTNEIKWSSR-----WDHVLKTMPQSNIQWFSILNSVIIVLFLSGMLAMILLKTLH 710 V++ T+ + S R +T+ I W SI+NS+++V L G +A+IL++ L Sbjct: 206 VRWSETSVERRSDRRRGDDGGFFPRTL---EIHWLSIINSMVLVFLLVGFVAVILMRVLR 262 Query: 711 RDIDRYNKLDTEEA---------QEEFGWKLVHGDVFR-PPRMGMTLCVFVGSGVQLILM 860 D+ RYN LD E Q + GWK++H DVFR PP G+ LC +G G Q + + Sbjct: 263 NDLARYN-LDEETTSAGSGDDFDQGDNGWKIIHTDVFRFPPYRGL-LCAVLGVGAQFLAL 320 Query: 861 TIVTLFISCFGFLSPANRGAFATCGIVFYVLFGAFAGFYSARLYKMMSGEKWKSNVLLTA 1040 + ++ G + GA + I+ Y L +G+ S+ Y+ + GE+W N++LT Sbjct: 321 GTGIIVMALLGMFNVHRHGAINSAAILLYALTCCISGYVSSHFYRQIGGERWVWNIILTT 380 Query: 1041 FLCPGSVFGIFFVLNIVLWASGSSAAVPFTTILGLLAMWFLISVPLCFVGAYFGFKK-QP 1217 L F + V+N V WA+GS+ A+P TTIL LL +W L+ PL +G FG P Sbjct: 381 SLFSVPFFLTWSVVNSVHWANGSTQALPATTILLLLTVWLLVGFPLTVIGGIFGKNNASP 440 Query: 1218 ISQPVRTNQIPRQIPYQYLYTRSIPSILMGGVLPFGCIFIQLFFILNSIWSTYVYYMFGX 1397 P RT I R+IP Q Y ++ + +GG LPF I ++L++I ++W Y ++G Sbjct: 441 FDAPCRTKNIAREIPPQPWYKSTVIHMTVGGFLPFSAISVELYYIFATVWGREQYTLYGI 500 Query: 1398 XXXXXXXXXXTCSETTILLCYFHLCAEDYRWWWRSFLTSGCTALYLFIYSIYFFHAKLDI 1577 + +I L YF L EDYRWWWRS L+ G T L++F+YS++++ + ++ Sbjct: 501 LFFVFAILLSVGACISIALTYFQLSGEDYRWWWRSVLSVGSTGLFIFLYSVFYYARRSNM 560 Query: 1578 KGSTSTFLYFGYTAIIIWLSFLVTGTVGFVSSFWFVRKIYSVVKVD 1715 G+ T +FGY+ + ++ FL+ GT+ F SS F+R IY +K+D Sbjct: 561 SGAVQTVEFFGYSLLTGYVFFLMLGTISFFSSLKFIRYIYVNLKMD 606
>sp|Q5R8F1|TM9S1_PONPY Transmembrane 9 superfamily protein member 1 precursor Length = 606 Score = 277 bits (709), Expect = 6e-74 Identities = 179/582 (30%), Positives = 294/582 (50%), Gaps = 14/582 (2%) Frame = +3 Query: 12 NLGQILFGERIQNSPYKIEFGKDLSYKEVCQKKYSKNDASQSKLKFLMSGIMLNYRHHWI 191 +LG++L G+R+ S Y+I F +++ + +C + S Q L I Y ++ Sbjct: 78 SLGEVLDGDRMAESLYEIRFRENVEKRILCHMQLSSAQVEQ-----LRQAIEELYYFEFV 132 Query: 192 VDDLPVVSCYKSGSNEVCYNGFPIGCYQSDKAKSVMCTTDASQNIPNTYFLFNHISLTIG 371 VDDLP+ GF +G + S +P+++ IG Sbjct: 133 VDDLPI-------------RGF-VGYMEE------------SGFLPHSH--------KIG 158 Query: 372 IYTD-EINEKLSKGRIVYVRVNTASKAFVKEGATDKELANAPPMILKGNEDTVIRYTYSV 548 ++T + + + RI++ V+ VK + D L+ +E + + YSV Sbjct: 159 LWTHLDFHLEFRGDRIIFANVSVRD---VKPHSLDG---------LRPDEFLGLTHAYSV 206 Query: 549 KFEPTN-EIKWSSRWDHVLKTMPQS-NIQWFSILNSVIIVLFLSGMLAMILLKTLHRDID 722 ++ T+ E + R P++ I W SI+NS+++V L G +A+IL++ L D+ Sbjct: 207 RWSETSVERRSDRRHGDDGGFFPRTLEIHWLSIINSMVLVFLLVGFVAVILMRVLRNDLA 266 Query: 723 RYNKLDTEEA---------QEEFGWKLVHGDVFR-PPRMGMTLCVFVGSGVQLILMTIVT 872 RYN LD E Q + GWK++H DVFR PP G+ LC +G G Q + + Sbjct: 267 RYN-LDEETTSAGSGDDFDQGDNGWKIIHTDVFRFPPYRGL-LCAVLGVGAQFLALGTGI 324 Query: 873 LFISCFGFLSPANRGAFATCGIVFYVLFGAFAGFYSARLYKMMSGEKWKSNVLLTAFLCP 1052 + ++ G + GA + I+ Y L +G+ S+ Y+ + GE+W N++LT L Sbjct: 325 IVMALLGMFNVHRHGAINSAAILLYALTCCISGYVSSHFYRQIGGERWVWNIILTTSLFS 384 Query: 1053 GSVFGIFFVLNIVLWASGSSAAVPFTTILGLLAMWFLISVPLCFVGAYFGFKK-QPISQP 1229 F + V+N V WA+GS+ A+P TTIL LL +W L+ PL +G FG P P Sbjct: 385 VPFFLTWSVVNSVHWANGSTQALPATTILLLLTVWLLVGFPLTVIGGIFGKNNASPFDAP 444 Query: 1230 VRTNQIPRQIPYQYLYTRSIPSILMGGVLPFGCIFIQLFFILNSIWSTYVYYMFGXXXXX 1409 RT I R+IP Q Y ++ + +GG LPF I ++L++I ++W Y ++G Sbjct: 445 CRTKNIAREIPPQPWYKSTVIHMTVGGFLPFSAISVELYYIFATVWGREQYTLYGILFFV 504 Query: 1410 XXXXXXTCSETTILLCYFHLCAEDYRWWWRSFLTSGCTALYLFIYSIYFFHAKLDIKGST 1589 + +I L YF L EDYRWWWRS L+ G T L++F+YS++++ + ++ G+ Sbjct: 505 FAILLSVGACISIALTYFQLSGEDYRWWWRSVLSVGSTGLFIFLYSVFYYARRSNMSGAV 564 Query: 1590 STFLYFGYTAIIIWLSFLVTGTVGFVSSFWFVRKIYSVVKVD 1715 T +FGY+ + ++ FL+ GT+ F SS F+R IY +K+D Sbjct: 565 QTVEFFGYSLLTGYVFFLMLGTISFFSSLKFIRYIYVNLKMD 606
>sp|Q9HD45|TM9S3_HUMAN Transmembrane 9 superfamily protein member 3 precursor (SM-11044 binding protein) (EP70-P-iso) Length = 589 Score = 268 bits (686), Expect = 3e-71 Identities = 176/571 (30%), Positives = 278/571 (48%), Gaps = 10/571 (1%) Frame = +3 Query: 33 GERIQNSPYKIEF-GKDLSYKE-VCQKKYSKNDASQSKLKFLMSGIMLNYRHHWIVDDLP 206 GE +Q ++EF G D+ +K+ V Y + D + K + I +Y + +DDLP Sbjct: 81 GEALQG--VELEFSGLDIKFKDDVMPATYCEIDLDKEKRDAFVYAIKNHYWYQMYIDDLP 138 Query: 207 VVSCYKSGSNEVCYNGFPIGCYQSDKAKSVMCTTDASQNIPNTYFLFNHISLTIGIYTDE 386 + E NG Y+L+ + L IG + Sbjct: 139 IWGIV----GEADENG-------------------------EDYYLWTYKKLEIGFNGNR 169 Query: 387 INEKLSKGRIVYVRVNTASKAFVKEGATDKELANAPPMILKGNEDTVIRYTYSVKFEPTN 566 I V VN S+ VK L N T I+ +YSVK++ + Sbjct: 170 I-----------VDVNLTSEGKVK---------------LVPN--TKIQMSYSVKWKKS- 200 Query: 567 EIKWSSRWDHVLK-TMPQSNIQWFSILNSVIIVLFLSGMLAMILLKTLHRDIDRYNKLDT 743 ++K+ R+D L + Q I WFSI NS ++V+FL G+++MIL++TL +D RY+K + Sbjct: 201 DVKFEDRFDKYLDPSFFQHRIHWFSIFNSFMMVIFLVGLVSMILMRTLRKDYARYSKEEE 260 Query: 744 -----EEAQEEFGWKLVHGDVFRPPRMGMTLCVFVGSGVQLILMTIVTLFISCFGFLSPA 908 + +E+GWK VHGDVFRP + +GSG Q+ ++++ + ++ L Sbjct: 261 MDDMDRDLGDEYGWKQVHGDVFRPSSHPLIFSSLIGSGCQIFAVSLIVIIVAMIEDLY-T 319 Query: 909 NRGAFATCGIVFYVLFGAFAGFYSARLYKMMSGEKWKSNVLLTAFLCPGSVFGIFFVLNI 1088 RG+ + I Y G++ LY G +W + + AFL P V G F +N Sbjct: 320 ERGSMLSTAIFVYAATSPVNGYFGGSLYARQGGRRWIKQMFIGAFLIPAMVCGTAFFINF 379 Query: 1089 VLWASGSSAAVPFTTILGLLAMWFLISVPLCFVGAYFG--FKKQPISQPVRTNQIPRQIP 1262 + +S A+PF T++ + + F + +PL VG G QP + P R N +PR IP Sbjct: 380 IAIYYHASRAIPFGTMVAVCCICFFVILPLNLVGTILGRNLSGQP-NFPCRVNAVPRPIP 438 Query: 1263 YQYLYTRSIPSILMGGVLPFGCIFIQLFFILNSIWSTYVYYMFGXXXXXXXXXXXTCSET 1442 + + + +GG+LPFG IFI+++FI S W+ +YY++G Sbjct: 439 EKKWFMEPAVIVCLGGILPFGSIFIEMYFIFTSFWAYKIYYVYGFMMLVLVILCIVTVCV 498 Query: 1443 TILLCYFHLCAEDYRWWWRSFLTSGCTALYLFIYSIYFFHAKLDIKGSTSTFLYFGYTAI 1622 TI+ YF L AEDYRW W SFL++ TA+Y+++YS Y++ K + G T YFGY A+ Sbjct: 499 TIVCTYFLLNAEDYRWQWTSFLSAASTAIYVYMYSFYYYFFKTKMYGLFQTSFYFGYMAV 558 Query: 1623 IIWLSFLVTGTVGFVSSFWFVRKIYSVVKVD 1715 ++ G +G++ + FVRKIY+ VK+D Sbjct: 559 FSTALGIMCGAIGYMGTSAFVRKIYTNVKID 589
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 212,966,118
Number of Sequences: 369166
Number of extensions: 4593418
Number of successful extensions: 11942
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 10946
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 11876
length of database: 68,354,980
effective HSP length: 117
effective length of database: 46,740,985
effective search space used: 22949823635
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail