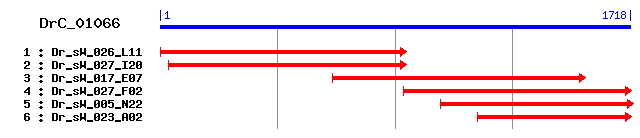
DrC_01066
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01066 (1717 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q99832|TCPH_HUMAN T-complex protein 1, eta subunit (TCP-... 764 0.0 sp|Q5R5C8|TCPH_PONPY T-complex protein 1, eta subunit (TCP-... 763 0.0 sp|P80313|TCPH_MOUSE T-complex protein 1, eta subunit (TCP-... 762 0.0 sp|P87153|TCPH_SCHPO Probable T-complex protein 1, eta subu... 647 0.0 sp|P54409|TCPH_TETPY T-complex protein 1, eta subunit (TCP-... 610 e-174 sp|P42943|TCPH_YEAST T-complex protein 1, eta subunit (TCP-... 603 e-172 sp|O77323|TCPH_PLAF7 T-complex protein 1, eta subunit (TCP-... 565 e-160 sp|P54410|TCPH_TETTH T-complex protein 1, eta subunit (TCP-... 430 e-120 sp|P50016|THS_METKA Thermosome subunit (Chaperonin-like com... 355 2e-97 sp|O28045|THSA_ARCFU Thermosome alpha subunit (Thermosome s... 355 2e-97
>sp|Q99832|TCPH_HUMAN T-complex protein 1, eta subunit (TCP-1-eta) (CCT-eta) (HIV-1 Nef interacting protein) Length = 543 Score = 764 bits (1973), Expect = 0.0 Identities = 383/543 (70%), Positives = 450/543 (82%) Frame = +2 Query: 26 MLPGAQIVLLKEGTDTSQGIPQIMSNITAIQSIAEAVRTTLGPRGMDKLIIDEKSKATIS 205 M+P ++LLKEGTD+SQGIPQ++SNI+A Q IAEAVRTTLGPRGMDKLI+D + KATIS Sbjct: 1 MMP-TPVILLKEGTDSSQGIPQLVSNISACQVIAEAVRTTLGPRGMDKLIVDGRGKATIS 59 Query: 206 NDGATILKLLDVVHPVASTVVQIALSQDDEVGDGTTSVMLLASEFLKQAKSFIEEGVHPE 385 NDGATILKLLDVVHP A T+V IA SQD EVGDGTTSV LLA+EFLKQ K ++EEG+HP+ Sbjct: 60 NDGATILKLLDVVHPAAKTLVDIAKSQDAEVGDGTTSVTLLAAEFLKQVKPYVEEGLHPQ 119 Query: 386 LIQXXXXXXXXXXXXXXXXXSVKIEKGNDAERRSLLEKCASTALSSKLVATYKEFFSKLV 565 +I +V ++K + E+R LLEKCA TALSSKL++ K FF+K+V Sbjct: 120 IIIRAFRTATQLAVNKIKEIAVTVKKADKVEQRKLLEKCAMTALSSKLISQQKAFFAKMV 179 Query: 566 VDAVSSLDHLLPLDMIGIKKVPGGSLQESQLIAGVAFKKTFSYAGFEMQPKSYKKPKISL 745 VDAV LD LL L MIGIKKV GG+L++SQL+AGVAFKKTFSYAGFEMQPK Y PKI+L Sbjct: 180 VDAVMMLDDLLQLKMIGIKKVQGGALEDSQLVAGVAFKKTFSYAGFEMQPKKYHNPKIAL 239 Query: 746 LNIELELKSEKENAEIRVSSVEEYQRIVDAEWQILYDKLEAIYKSGAKIVLSKLPIGDVA 925 LN+ELELK+EK+NAEIRV +VE+YQ IVDAEW ILYDKLE I+ SGAK+VLSKLPIGDVA Sbjct: 240 LNVELELKAEKDNAEIRVHTVEDYQAIVDAEWNILYDKLEKIHHSGAKVVLSKLPIGDVA 299 Query: 926 TQYFADRDMFCAGRVTEEDLKRTMAACGGAIQTTVKDITDDTLGKCESFEEVQIGGERFN 1105 TQYFADRDMFCAGRV EEDLKRTM ACGG+IQT+V ++ D LG+C+ FEE QIGGER+N Sbjct: 300 TQYFADRDMFCAGRVPEEDLKRTMMACGGSIQTSVNALSADVLGRCQVFEETQIGGERYN 359 Query: 1106 IFVGCPEAKTCTIILRGGSEQFMEETERSLHDAIMIVRRAMKNDAVVAGGGAIDMEISKY 1285 F GCP+AKTCT ILRGG+EQFMEETERSLHDAIMIVRRA+KND+VVAGGGAI+ME+SKY Sbjct: 360 FFTGCPKAKTCTFILRGGAEQFMEETERSLHDAIMIVRRAIKNDSVVAGGGAIEMELSKY 419 Query: 1286 LRDYSRNIHGRDQQFIGAMARAFEVIPRQLCENAGFDATNILNKLRQKHAQGKIWCGVDI 1465 LRDYSR I G+ Q IGA A+A E+IPRQLC+NAGFDATNILNKLR +HAQG W GVDI Sbjct: 420 LRDYSRTIPGKQQLLIGAYAKALEIIPRQLCDNAGFDATNILNKLRARHAQGGTWYGVDI 479 Query: 1466 MNEDIADNFEACVWEPALVKSNAITAATEAACMVLTLDETIRNAKSKTPEDNRPVKMPGR 1645 NEDIADNFEA VWEPA+V+ NA+TAA+EAAC+++++DETI+N +S + GR Sbjct: 480 NNEDIADNFEAFVWEPAMVRINALTAASEAACLIVSVDETIKNPRSTVDAPTAAGRGRGR 539 Query: 1646 GRP 1654 GRP Sbjct: 540 GRP 542
>sp|Q5R5C8|TCPH_PONPY T-complex protein 1, eta subunit (TCP-1-eta) (CCT-eta) Length = 543 Score = 763 bits (1970), Expect = 0.0 Identities = 382/543 (70%), Positives = 450/543 (82%) Frame = +2 Query: 26 MLPGAQIVLLKEGTDTSQGIPQIMSNITAIQSIAEAVRTTLGPRGMDKLIIDEKSKATIS 205 M+P ++LLKEGTD+SQGIPQ++SNI+A Q IAEAVRTTLGPRGMDKLI+D + KATIS Sbjct: 1 MMP-TPVILLKEGTDSSQGIPQLVSNISACQVIAEAVRTTLGPRGMDKLIVDGRGKATIS 59 Query: 206 NDGATILKLLDVVHPVASTVVQIALSQDDEVGDGTTSVMLLASEFLKQAKSFIEEGVHPE 385 NDGATILKLLDVVHP A T+V IA SQD EVGDGTTSV LLA+EFLKQ K ++EEG+HP+ Sbjct: 60 NDGATILKLLDVVHPAAKTLVDIAKSQDAEVGDGTTSVTLLAAEFLKQVKPYVEEGLHPQ 119 Query: 386 LIQXXXXXXXXXXXXXXXXXSVKIEKGNDAERRSLLEKCASTALSSKLVATYKEFFSKLV 565 +I +V ++K + E+R LLEKCA TALSSKL++ K FF+K+V Sbjct: 120 IIIRAFRTATQLAVNKIKEIAVTVKKADKVEQRKLLEKCAMTALSSKLISQQKAFFAKMV 179 Query: 566 VDAVSSLDHLLPLDMIGIKKVPGGSLQESQLIAGVAFKKTFSYAGFEMQPKSYKKPKISL 745 VDAV LD LL L MIGIKKV GG+L++SQL+AGVAFKKTFSYAGFEMQPK Y PK++L Sbjct: 180 VDAVMMLDDLLQLKMIGIKKVQGGALEDSQLVAGVAFKKTFSYAGFEMQPKKYHNPKVAL 239 Query: 746 LNIELELKSEKENAEIRVSSVEEYQRIVDAEWQILYDKLEAIYKSGAKIVLSKLPIGDVA 925 LN+ELELK+EK+NAEIRV +VE+YQ IVDAEW ILYDKLE I+ SGAK+VLSKLPIGDVA Sbjct: 240 LNVELELKAEKDNAEIRVHTVEDYQAIVDAEWNILYDKLEKIHHSGAKVVLSKLPIGDVA 299 Query: 926 TQYFADRDMFCAGRVTEEDLKRTMAACGGAIQTTVKDITDDTLGKCESFEEVQIGGERFN 1105 TQYFADRDMFCAGRV EEDLKRTM ACGG+IQT+V ++ D LG+C+ FEE QIGGER+N Sbjct: 300 TQYFADRDMFCAGRVPEEDLKRTMMACGGSIQTSVNALSADVLGRCQVFEETQIGGERYN 359 Query: 1106 IFVGCPEAKTCTIILRGGSEQFMEETERSLHDAIMIVRRAMKNDAVVAGGGAIDMEISKY 1285 F GCP+AKTCT ILRGG+EQFMEETERSLHDAIMIVRRA+KND+VVAGGGAI+ME+SKY Sbjct: 360 FFTGCPKAKTCTFILRGGAEQFMEETERSLHDAIMIVRRAIKNDSVVAGGGAIEMELSKY 419 Query: 1286 LRDYSRNIHGRDQQFIGAMARAFEVIPRQLCENAGFDATNILNKLRQKHAQGKIWCGVDI 1465 LRDYSR I G+ Q IGA A+A E+IPRQLC+NAGFDATNILNKLR +HAQG W GVDI Sbjct: 420 LRDYSRTIPGKQQLLIGAYAKALEIIPRQLCDNAGFDATNILNKLRARHAQGGTWYGVDI 479 Query: 1466 MNEDIADNFEACVWEPALVKSNAITAATEAACMVLTLDETIRNAKSKTPEDNRPVKMPGR 1645 NEDIADNFEA VWEPA+V+ NA+TAA+EAAC+++++DETI+N +S + GR Sbjct: 480 NNEDIADNFEAFVWEPAMVRINALTAASEAACLIVSVDETIKNPRSTVDAPAAAGRGRGR 539 Query: 1646 GRP 1654 GRP Sbjct: 540 GRP 542
>sp|P80313|TCPH_MOUSE T-complex protein 1, eta subunit (TCP-1-eta) (CCT-eta) Length = 544 Score = 762 bits (1967), Expect = 0.0 Identities = 382/542 (70%), Positives = 452/542 (83%) Frame = +2 Query: 26 MLPGAQIVLLKEGTDTSQGIPQIMSNITAIQSIAEAVRTTLGPRGMDKLIIDEKSKATIS 205 M+P ++LLKEGTD+SQGIPQ++SNI+A Q IAEAVRTTLGPRGMDKLI+D + KATIS Sbjct: 1 MMP-TPVILLKEGTDSSQGIPQLVSNISACQVIAEAVRTTLGPRGMDKLIVDGRGKATIS 59 Query: 206 NDGATILKLLDVVHPVASTVVQIALSQDDEVGDGTTSVMLLASEFLKQAKSFIEEGVHPE 385 NDGATILKLLDVVHP A T+V IA SQD EVGDGTTSV LLA+EFLKQ K ++EEG+HP+ Sbjct: 60 NDGATILKLLDVVHPAAKTLVDIAKSQDAEVGDGTTSVTLLAAEFLKQVKPYVEEGLHPQ 119 Query: 386 LIQXXXXXXXXXXXXXXXXXSVKIEKGNDAERRSLLEKCASTALSSKLVATYKEFFSKLV 565 +I +V ++K + E+R +LEKCA TALSSKL++ K FF+K+V Sbjct: 120 IIIRAFRTATQLAVNKIKEIAVTVKKQDKVEQRKMLEKCAMTALSSKLISQQKVFFAKMV 179 Query: 566 VDAVSSLDHLLPLDMIGIKKVPGGSLQESQLIAGVAFKKTFSYAGFEMQPKSYKKPKISL 745 VDAV LD LL L MIGIKKV GG+L+ESQL+AGVAFKKTFSYAGFEMQPK YK PKI+L Sbjct: 180 VDAVMMLDELLQLKMIGIKKVQGGALEESQLVAGVAFKKTFSYAGFEMQPKKYKNPKIAL 239 Query: 746 LNIELELKSEKENAEIRVSSVEEYQRIVDAEWQILYDKLEAIYKSGAKIVLSKLPIGDVA 925 LN+ELELK+EK+NAEIRV +VE+YQ IVDAEW ILYDKLE I++SGAK++LSKLPIGDVA Sbjct: 240 LNVELELKAEKDNAEIRVHTVEDYQAIVDAEWNILYDKLEKIHQSGAKVILSKLPIGDVA 299 Query: 926 TQYFADRDMFCAGRVTEEDLKRTMAACGGAIQTTVKDITDDTLGKCESFEEVQIGGERFN 1105 TQYFADRDMFCAGRV EEDLKRTM ACGG+IQT+V + D LG C+ FEE QIGGER+N Sbjct: 300 TQYFADRDMFCAGRVPEEDLKRTMMACGGSIQTSVNALVPDVLGHCQVFEETQIGGERYN 359 Query: 1106 IFVGCPEAKTCTIILRGGSEQFMEETERSLHDAIMIVRRAMKNDAVVAGGGAIDMEISKY 1285 F GCP+AKTCTIILRGG+EQFMEETERSLHDAIMIVRRA+KND+VVAGGGAI+ME+SKY Sbjct: 360 FFTGCPKAKTCTIILRGGAEQFMEETERSLHDAIMIVRRAIKNDSVVAGGGAIEMELSKY 419 Query: 1286 LRDYSRNIHGRDQQFIGAMARAFEVIPRQLCENAGFDATNILNKLRQKHAQGKIWCGVDI 1465 LRDYSR I G+ Q IGA A+A E+IPRQLC+NAGFDATNILNKLR +HAQG +W GVDI Sbjct: 420 LRDYSRTIPGKQQLLIGAYAKALEIIPRQLCDNAGFDATNILNKLRARHAQGGMWYGVDI 479 Query: 1466 MNEDIADNFEACVWEPALVKSNAITAATEAACMVLTLDETIRNAKSKTPEDNRPVKMPGR 1645 NE+IADNF+A VWEPA+V+ NA+TAA+EAAC+++++DETI+N +S + P GR Sbjct: 480 NNENIADNFQAFVWEPAMVRINALTAASEAACLIVSVDETIKNPRSTV---DPPAPSAGR 536 Query: 1646 GR 1651 GR Sbjct: 537 GR 538
>sp|P87153|TCPH_SCHPO Probable T-complex protein 1, eta subunit (TCP-1-eta) (CCT-eta) Length = 558 Score = 647 bits (1668), Expect = 0.0 Identities = 320/552 (57%), Positives = 423/552 (76%), Gaps = 11/552 (1%) Frame = +2 Query: 32 PGAQIVLLKEGTDTSQGIPQIMSNITAIQSIAEAVRTTLGPRGMDKLIIDEKSKATISND 211 P +++LKEGTD SQG Q++SNI A ++ + +RTTLGP G DKL++D++ + ISND Sbjct: 6 PQIPVIVLKEGTDDSQGRGQLLSNINACVAVQDTIRTTLGPLGADKLMVDDRGEVVISND 65 Query: 212 GATILKLLDVVHPVASTVVQIALSQDDEVGDGTTSVMLLASEFLKQAKSFIEEGVHPELI 391 GATI+KLLD+VHP A T+V IA +QD EVGDGTTSV++ A E L++A++F+E+GV LI Sbjct: 66 GATIMKLLDIVHPAAKTLVDIARAQDAEVGDGTTSVVVFAGELLREARTFVEDGVSSHLI 125 Query: 392 QXXXXXXXXXXXXXXXXXSVKIEKGNDAERRSLLEKCASTALSSKLVATYKEFFSKLVVD 571 ++ ++ ++ + R LL KCASTA++SKL+ + FF+K+VVD Sbjct: 126 IRGYRKAAQLAVNKIKEIAIHLDLSDEGKLRDLLTKCASTAMNSKLIRSNSTFFTKMVVD 185 Query: 572 AVSSLDHL-LPLDMIGIKKVPGGSLQESQLIAGVAFKKTFSYAGFEMQPKSYKKPKISLL 748 AV +LD L +MIGIKKVPGG++++S L+ GVAFKKTFSYAGFE QPK +K PKI L Sbjct: 186 AVLTLDQEDLNENMIGIKKVPGGAMEDSLLVKGVAFKKTFSYAGFEQQPKFFKNPKILCL 245 Query: 749 NIELELKSEKENAEIRVSSVEEYQRIVDAEWQILYDKLEAIYKSGAKIVLSKLPIGDVAT 928 ++ELELK+EK+NAE+RV V+EYQ IVDAEW+I++ KLEAI +GAK+VLSKLPIGD+AT Sbjct: 246 DVELELKAEKDNAEVRVDKVQEYQNIVDAEWRIIFSKLEAIVATGAKVVLSKLPIGDLAT 305 Query: 929 QYFADRDMFCAGRVTEEDLKRTMAACGGAIQTTVKDITDDTLGKCESFEEVQIGGERFNI 1108 QYFADRD+FCAGRV +DL R + A GG+IQ+T +I + LG C++FEE QIGG+RFN+ Sbjct: 306 QYFADRDIFCAGRVAADDLNRVVQAVGGSIQSTCSNIEEKHLGTCDTFEERQIGGDRFNL 365 Query: 1109 FVGCPEAKTCTIILRGGSEQFMEETERSLHDAIMIVRRAMKNDAVVAGGGAIDMEISKYL 1288 F GCP+AKTCT+ILRGG++QF+ E ERSLHDAIMIV+ A+KN+ VVAGGGA +ME+SKYL Sbjct: 366 FEGCPKAKTCTLILRGGADQFIAEVERSLHDAIMIVKHALKNNLVVAGGGACEMELSKYL 425 Query: 1289 RDYSRNIHGRDQQFIGAMARAFEVIPRQLCENAGFDATNILNKLRQKHAQGKIWCGVDIM 1468 RDYS I G+ Q FI A AR+ EVIPRQLC+NAGFD+TNILNKLR +HA+G++W GVD+ Sbjct: 426 RDYSLTISGKQQNFIAAFARSLEVIPRQLCDNAGFDSTNILNKLRMQHAKGEMWAGVDMD 485 Query: 1469 NEDIADNFEACVWEPALVKSNAITAATEAACMVLTLDETIRNAKSKTPE----------D 1618 +E +A+NFE VWEP+ VKSNAI +ATEAA ++L++DETI+N S+ P+ Sbjct: 486 SEGVANNFEKFVWEPSTVKSNAILSATEAATLILSVDETIKNEPSQQPQAPQGALPPGAA 545 Query: 1619 NRPVKMPGRGRP 1654 NR ++ GRG P Sbjct: 546 NRIMRGRGRGMP 557
>sp|P54409|TCPH_TETPY T-complex protein 1, eta subunit (TCP-1-eta) (CCT-eta) Length = 558 Score = 610 bits (1572), Expect = e-174 Identities = 318/530 (60%), Positives = 400/530 (75%), Gaps = 6/530 (1%) Frame = +2 Query: 44 IVLLKEGTDTSQGIPQIMSNITAIQSIAEAVRTTLGPRGMDKLIIDEKSKATISNDGATI 223 I+LLK+GTDTSQG QI+SNI A+QSI E V+TTLGPRGMDKLI + ATISNDGATI Sbjct: 6 ILLLKDGTDTSQGKAQIISNINAVQSIVEIVKTTLGPRGMDKLIEGNRG-ATISNDGATI 64 Query: 224 LKLLDVVHPVASTVVQIALSQDDEVGDGTTSVMLLASEFLKQAKSFIEEGVHPELIQXXX 403 L LLD+VHP A T+V IA +QDDEVGDGTTSV LLA E LK++K+FIEEG+HP+++ Sbjct: 65 LNLLDIVHPAAKTLVDIAKAQDDEVGDGTTSVCLLAGELLKESKNFIEEGMHPQIVTKGY 124 Query: 404 XXXXXXXXXXXXXXSVKIEKGNDAERRSLLEKCASTALSSKLVATYKEFFSKLVVDAVSS 583 S + +D E+R +L KCA T+L+SKL+A YKEFFS++VV AV + Sbjct: 125 KEALKLALTFLQENSYSVADKSDGEKREMLLKCAQTSLNSKLLAHYKEFFSEMVVQAVET 184 Query: 584 LD-HLLPLDMIGIKKVPGGSLQESQLIAGVAFKKTFSYAGFEMQPKSYKKPKISLLNIEL 760 LD +LL D+IGIK V GGS+ +S L+ GVAFKKTFSYAGFE QPK + PKI LLNIEL Sbjct: 185 LDTNLLDKDLIGIKMVTGGSVTDSVLVKGVAFKKTFSYAGFEQQPKKFANPKICLLNIEL 244 Query: 761 ELKSEKENAEIRVSSVEEYQRIVDAEWQILYDKLEAIYKSGAKIVLSKLPIGDVATQYFA 940 ELK+EKENAEIR+ + ++Y+ IVDAEW+++Y+KL I +SGA+IVLSKLPIGD+ATQYFA Sbjct: 245 ELKAEKENAEIRIDNPDDYKSIVDAEWELIYEKLRKIVESGAQIVLSKLPIGDLATQYFA 304 Query: 941 DRDMFCAGRVTEEDLKRTMAACGGAIQTTVKDITDDTLGKCESFEEVQIGGERFNIFVGC 1120 DR++FCAGRV ED+KR A G +QTTV ++ D LG C FEE QIG ER+N+F C Sbjct: 305 DRNIFCAGRVDAEDIKRVQKATGSIVQTTVNGLSQDVLGTCGMFEEQQIGAERYNLFQDC 364 Query: 1121 PEAKTCTIILRGGSEQFMEETERSLHDAIMIVRRAMKNDAVVAGGGAIDMEISKYLRDYS 1300 P +K+ TIILRGG+EQF+ E ERSL+DAIMIVRR MK + +V GGGAI++EIS+ LR +S Sbjct: 365 PHSKSATIILRGGAEQFIAEAERSLNDAIMIVRRCMKANKIVPGGGAIELEISRLLRLHS 424 Query: 1301 RNIHGRDQQFIGAMARAFEVIPRQLCENAGFDATNILNKLRQKHA----QGKIWCGVDIM 1468 R G+ Q I A A+A EVIP+ + +NAG D+ +LNKLRQKHA Q K + GVDI Sbjct: 425 RKTEGKVQLVINAFAKALEVIPKTIADNAGHDSIQVLNKLRQKHALESDQSKNF-GVDIN 483 Query: 1469 NED-IADNFEACVWEPALVKSNAITAATEAACMVLTLDETIRNAKSKTPE 1615 D I +NFE VWEP +V+ NA +AATEAAC +L++DET+RN KS+ P+ Sbjct: 484 AVDGIGNNFENFVWEPIIVRKNAFSAATEAACTILSIDETVRNPKSEQPK 533
>sp|P42943|TCPH_YEAST T-complex protein 1, eta subunit (TCP-1-eta) (CCT-eta) Length = 550 Score = 603 bits (1556), Expect = e-172 Identities = 312/538 (57%), Positives = 393/538 (73%), Gaps = 2/538 (0%) Frame = +2 Query: 44 IVLLKEGTDTSQGIPQIMSNITAIQSIAEAVRTTLGPRGMDKLIIDEKSKATISNDGATI 223 IV+LKEGTD SQG QI+SNI A ++ EA++ TLGP G D LI+ K TISNDGATI Sbjct: 10 IVVLKEGTDASQGKGQIISNINACVAVQEALKPTLGPLGSDILIVTSNQKTTISNDGATI 69 Query: 224 LKLLDVVHPVASTVVQIALSQDDEVGDGTTSVMLLASEFLKQAKSFIEEGVHPELIQXXX 403 LKLLDVVHP A T+V I+ +QD EVGDGTTSV +LA E +K+AK F+EEG+ LI Sbjct: 70 LKLLDVVHPAAKTLVDISRAQDAEVGDGTTSVTILAGELMKEAKPFLEEGISSHLIMKGY 129 Query: 404 XXXXXXXXXXXXXXSVKIEKGNDAERRSLLEKCASTALSSKLVATYKEFFSKLVVDAVSS 583 +V I + R LLE+CA TA+SSKL+ +FF K+ VDAV S Sbjct: 130 RKAVSLAVEKINELAVDITS-EKSSGRELLERCARTAMSSKLIHNNADFFVKMCVDAVLS 188 Query: 584 LDHL-LPLDMIGIKKVPGGSLQESQLIAGVAFKKTFSYAGFEMQPKSYKKPKISLLNIEL 760 LD L +IGIKK+PGG+++ES I GVAFKKTFSYAGFE QPK + PKI LN+EL Sbjct: 189 LDRNDLDDKLIGIKKIPGGAMEESLFINGVAFKKTFSYAGFEQQPKKFNNPKILSLNVEL 248 Query: 761 ELKSEKENAEIRVSSVEEYQRIVDAEWQILYDKLEAIYKSGAKIVLSKLPIGDVATQYFA 940 ELK+EK+NAE+RV VE+YQ IVDAEWQ++++KL + ++GA IVLSKLPIGD+ATQ+FA Sbjct: 249 ELKAEKDNAEVRVEHVEDYQAIVDAEWQLIFEKLRQVEETGANIVLSKLPIGDLATQFFA 308 Query: 941 DRDMFCAGRVTEEDLKRTMAACGGAIQTTVKDITDDTLGKCESFEEVQIGGERFNIFVGC 1120 DR++FCAGRV+ +D+ R + A GG+IQ+T DI + LG C FEE+QIG ER+N+F GC Sbjct: 309 DRNIFCAGRVSADDMNRVIQAVGGSIQSTTSDIKPEHLGTCALFEEMQIGSERYNLFQGC 368 Query: 1121 PEAKTCTIILRGGSEQFMEETERSLHDAIMIVRRAMKNDAVVAGGGAIDMEISKYLRDYS 1300 P+AKTCT++LRGG+EQ + E ERSLHDAIMIV+RA++N +VAGGGA +ME+SK LRDYS Sbjct: 369 PQAKTCTLLLRGGAEQVIAEVERSLHDAIMIVKRALQNKLIVAGGGATEMEVSKCLRDYS 428 Query: 1301 RNIHGRDQQFIGAMARAFEVIPRQLCENAGFDATNILNKLRQKHAQGKIWCGVDIMNEDI 1480 + I G+ Q I A A+A EVIPRQLCENAGFDA ILNKLR H++G+ W GV E+I Sbjct: 429 KTIAGKQQMIINAFAKALEVIPRQLCENAGFDAIEILNKLRLAHSKGEKWYGVVFETENI 488 Query: 1481 ADNFEACVWEPALVKSNAITAATEAACMVLTLDETIRNAKSKTPEDN-RPVKMPGRGR 1651 DNF VWEPALVK NA+ +ATEA ++L++DETI N S++ P + GRGR Sbjct: 489 GDNFAKFVWEPALVKINALNSATEATNLILSVDETITNKGSESANAGMMPPQGAGRGR 546
>sp|O77323|TCPH_PLAF7 T-complex protein 1, eta subunit (TCP-1-eta) (CCT-eta) Length = 539 Score = 565 bits (1456), Expect = e-160 Identities = 292/530 (55%), Positives = 377/530 (71%), Gaps = 2/530 (0%) Frame = +2 Query: 44 IVLLKEGTDTSQGIPQIMSNITAIQSIAEAVRTTLGPRGMDKLIIDEKSKATISNDGATI 223 IVLLKEGTDT+QG QI+ NI A Q I + V+TTLGPRGMDKLI E+ TI+NDGAT+ Sbjct: 9 IVLLKEGTDTAQGRSQIIRNINACQIIVDIVKTTLGPRGMDKLIYTERD-VTITNDGATV 67 Query: 224 LKLLDVVHPVASTVVQIALSQDDEVGDGTTSVMLLASEFLKQAKSFIEEGVHPELIQXXX 403 + LL++ HP AS +V IA SQDDEVGDGTTSV+++A E L +AK + +G+ P +I Sbjct: 68 MNLLNISHPAASILVDIAKSQDDEVGDGTTSVVVVAGELLNEAKGLLNDGIEPNMIIDGF 127 Query: 404 XXXXXXXXXXXXXXSVKIEKGNDAERRSLLEKCASTALSSKLVATYKEFFSKLVVDAVSS 583 S+ N+ E+RS+L KCA TAL+SKLV+ +KEFF +LVV+AV Sbjct: 128 RNACNVAINKLNELSLNFSNKNEEEKRSILLKCAQTALNSKLVSNHKEFFGELVVNAVYK 187 Query: 584 LDHLLPLDMIGIKKVPGGSLQESQLIAGVAFKKTFSYAGFEMQPKSYKKPKISLLNIELE 763 L L IGIKKV GGS ++QLI GVAFKKTFSYAGFE QPK + PKI LLN+ELE Sbjct: 188 LGDNLDKSNIGIKKVTGGSCLDTQLIYGVAFKKTFSYAGFEQQPKKFINPKILLLNVELE 247 Query: 764 LKSEKENAEIRVSSVEEYQRIVDAEWQILYDKLEAIYKSGAKIVLSKLPIGDVATQYFAD 943 LK+EKENAE+R+ + EY IV AEW I++ KL I GA IVLSKLPIGD+ATQ+FAD Sbjct: 248 LKAEKENAEVRIENPNEYNSIVQAEWDIIFKKLNLIKDCGANIVLSKLPIGDIATQFFAD 307 Query: 944 RDMFCAGRVTEEDLKRTMAACGGAIQTTVKDITDDTLGKCESFEEVQIGGERFNIFVGCP 1123 D+FCAGRV + DLKRT A G +QT++ ++ DD LG C FEEVQIG ER+NIF C Sbjct: 308 HDIFCAGRVEDADLKRTANATGALVQTSLFNLNDDVLGTCGVFEEVQIGNERYNIFKECL 367 Query: 1124 EAKTCTIILRGGSEQFMEETERSLHDAIMIVRRAMKNDAVVAGGGAIDMEISKYLRDYSR 1303 + K+ TIILRGG++QF+EE ERS++DAIMIV R + N +V G G+I+M++SKYLR YSR Sbjct: 368 KTKSVTIILRGGAKQFIEEVERSINDAIMIVLRCITNSEIVPGAGSIEMQLSKYLRIYSR 427 Query: 1304 NIHGRDQQFIGAMARAFEVIPRQLCENAGFDATNILNKLRQKHAQ--GKIWCGVDIMNED 1477 +I ++Q + + A+A E IPR L NAG+D+T+ILNKLR+KH++ IW GVD M D Sbjct: 428 SICNKEQIVLFSFAKALESIPRHLSHNAGYDSTDILNKLRKKHSEQTSDIWYGVDCMEGD 487 Query: 1478 IADNFEACVWEPALVKSNAITAATEAACMVLTLDETIRNAKSKTPEDNRP 1627 I + ++ C++E +K N I +ATEAAC++L++DETI+N S P Sbjct: 488 IINAYDNCIFEVTKIKRNVIYSATEAACLILSIDETIKNPSSAAGTQRSP 537
>sp|P54410|TCPH_TETTH T-complex protein 1, eta subunit (TCP-1-eta) (CCT-eta) Length = 353 Score = 430 bits (1106), Expect = e-120 Identities = 216/353 (61%), Positives = 270/353 (76%), Gaps = 1/353 (0%) Frame = +2 Query: 206 NDGATILKLLDVVHPVASTVVQIALSQDDEVGDGTTSVMLLASEFLKQAKSFIEEGVHPE 385 NDGATIL LLD+VHP A T+V IA +QDDEVGDGTTSV LLA E LK++K FIEEG+HP+ Sbjct: 1 NDGATILNLLDIVHPAAKTLVDIAKAQDDEVGDGTTSVCLLAGELLKESKYFIEEGMHPQ 60 Query: 386 LIQXXXXXXXXXXXXXXXXXSVKIEKGNDAERRSLLEKCASTALSSKLVATYKEFFSKLV 565 +I + + N+ E+R +L KCA T+L+SKL+A YKEFFS+LV Sbjct: 61 IITKGYKEALKLALQFLHENAHSVADKNETEKREMLIKCAQTSLNSKLLAHYKEFFSELV 120 Query: 566 VDAVSSLD-HLLPLDMIGIKKVPGGSLQESQLIAGVAFKKTFSYAGFEMQPKSYKKPKIS 742 V AV +L+ +LL D+IGIK V GGS+ +S L++GVAFKKTFSYAGFE QPK + PKI Sbjct: 121 VQAVETLETNLLDKDLIGIKMVTGGSVTDSFLVSGVAFKKTFSYAGFEQQPKKFNNPKIC 180 Query: 743 LLNIELELKSEKENAEIRVSSVEEYQRIVDAEWQILYDKLEAIYKSGAKIVLSKLPIGDV 922 LLNIELELK+EKENAEIR+ + ++Y+ IVDAEW+++Y+KL I +SGA IVLSKLPIGD+ Sbjct: 181 LLNIELELKAEKENAEIRIENPDDYKSIVDAEWELIYEKLRKIVESGANIVLSKLPIGDL 240 Query: 923 ATQYFADRDMFCAGRVTEEDLKRTMAACGGAIQTTVKDITDDTLGKCESFEEVQIGGERF 1102 ATQYFADR++FCAGRV ED+KR + G +QTTV +T+D LG C FEEVQIG ER+ Sbjct: 241 ATQYFADRNIFCAGRVDAEDMKRVQKSTGAIVQTTVNGLTEDVLGTCNQFEEVQIGAERY 300 Query: 1103 NIFVGCPEAKTCTIILRGGSEQFMEETERSLHDAIMIVRRAMKNDAVVAGGGA 1261 N+F CP +K+ TIILRGG+EQF+ E ERSL+DAIMIVRR MK + +V GGGA Sbjct: 301 NLFKDCPHSKSATIILRGGAEQFIAEAERSLNDAIMIVRRCMKANKIVPGGGA 353
>sp|P50016|THS_METKA Thermosome subunit (Chaperonin-like complex) (CLIC) Length = 545 Score = 355 bits (912), Expect = 2e-97 Identities = 186/524 (35%), Positives = 314/524 (59%), Gaps = 6/524 (1%) Frame = +2 Query: 35 GAQIVLLKEGTDTSQGIPQIMSNITAIQSIAEAVRTTLGPRGMDKLIIDEKSKATISNDG 214 G Q+++L EG G NI A + +AE VRTTLGP GMDK+++DE ++NDG Sbjct: 8 GRQVLILPEGYQRFVGRDAQRMNIMAARVVAETVRTTLGPMGMDKMLVDEMGDVVVTNDG 67 Query: 215 ATILKLLDVVHPVASTVVQIALSQDDEVGDGTTSVMLLASEFLKQAKSFIEEGVHPELIQ 394 TIL+ +D+ HP A VV++A +Q+DEVGDGTT+ ++LA E L +A+ +++ +HP +I Sbjct: 68 VTILEEMDIEHPAAKMVVEVAKTQEDEVGDGTTTAVVLAGELLHKAEDLLQQDIHPTVIA 127 Query: 395 XXXXXXXXXXXXXXXXXSVKIEKGNDAERRSLLEKCASTALSSKLVATYKEFFSKLVVDA 574 + +I D + L+K A TA++ K V +++ ++LVV A Sbjct: 128 RGYRMAVEKAEEILEEIAEEI----DPDDEETLKKIAKTAMTGKGVEKARDYLAELVVKA 183 Query: 575 VSSL------DHLLPLDMIGIKKVPGGSLQESQLIAGVAFKKTFSYAGFEMQPKSYKKPK 736 V + + ++ D I ++K GG L++++L+ G+ K + G P+ + K Sbjct: 184 VKQVAEEEDGEIVIDTDHIKLEKKEGGGLEDTELVKGMVIDKERVHPGM---PRRVENAK 240 Query: 737 ISLLNIELELKSEKENAEIRVSSVEEYQRIVDAEWQILYDKLEAIYKSGAKIVLSKLPIG 916 I+LLN +E+K + +AEIR++ E+ Q ++ E ++L + ++ I ++GA +V + I Sbjct: 241 IALLNCPIEVKETETDAEIRITDPEQLQAFIEEEERMLSEMVDKIAETGANVVFCQKGID 300 Query: 917 DVATQYFADRDMFCAGRVTEEDLKRTMAACGGAIQTTVKDITDDTLGKCESFEEVQIGGE 1096 D+A Y A + + RV + D+++ A G I T + D++++ LG+ E EE ++ G+ Sbjct: 301 DLAQHYLAKKGILAVRRVKKSDMQKLARATGARIVTNIDDLSEEDLGEAEVVEEKKVAGD 360 Query: 1097 RFNIFVGCPEAKTCTIILRGGSEQFMEETERSLHDAIMIVRRAMKNDAVVAGGGAIDMEI 1276 + GC + K TI++RGG+E ++E ER++ DAI +V A+++ VVAGGGA ++E+ Sbjct: 361 KMIFVEGCKDPKAVTILIRGGTEHVVDEAERAIEDAIGVVAAALEDGKVVAGGGAPEVEV 420 Query: 1277 SKYLRDYSRNIHGRDQQFIGAMARAFEVIPRQLCENAGFDATNILNKLRQKHAQGKIWCG 1456 ++ LRD++ + GR+Q + A A A E+IPR L EN+G D ++L +LR KH G++ G Sbjct: 421 ARQLRDFADGVEGREQLAVEAFADALEIIPRTLAENSGLDPIDVLVQLRAKHEDGQVTAG 480 Query: 1457 VDIMNEDIADNFEACVWEPALVKSNAITAATEAACMVLTLDETI 1588 +D+ + D+ D E V EP VK+ A+ +ATEAA M+L +D+ I Sbjct: 481 IDVYDGDVKDMLEEGVVEPLRVKTQALASATEAAEMILRIDDVI 524
>sp|O28045|THSA_ARCFU Thermosome alpha subunit (Thermosome subunit 1) (Chaperonin alpha subunit) Length = 545 Score = 355 bits (912), Expect = 2e-97 Identities = 200/538 (37%), Positives = 316/538 (58%), Gaps = 9/538 (1%) Frame = +2 Query: 29 LPGAQIVLLKEGTDTSQGIPQIMSNITAIQSIAEAVRTTLGPRGMDKLIIDEKSKATISN 208 L G +++LKEGT + G NI A + IAEAV++TLGP+GMDK+++D I+N Sbjct: 4 LQGTPVLILKEGTQRTVGRDAQRMNIMAARVIAEAVKSTLGPKGMDKMLVDSLGDVVITN 63 Query: 209 DGATILKLLDVVHPVASTVVQIALSQDDEVGDGTTSVMLLASEFLKQAKSFIEEGVHPEL 388 DG TILK +DV HP A ++++A +QD+EVGDGTT+ ++LA E LK+A+ +++ +HP + Sbjct: 64 DGVTILKEMDVEHPAAKMIIEVAKTQDNEVGDGTTTAVVLAGELLKKAEELLDQDIHPTV 123 Query: 389 IQXXXXXXXXXXXXXXXXXSVKIEKGNDAERRSLLEKCASTALSSKLVATYKEFFSKLVV 568 I ++ I D E L+K A+TA++ K + S LVV Sbjct: 124 IARGYRMAANKAVEILESIAMDI----DVEDEETLKKIAATAITGKHSEYALDHLSSLVV 179 Query: 569 DAVSSLD------HLLPLDMIGIKKVPGGSLQESQLIAGVAFKKTFSYAGFEMQPKSYKK 730 +AV + + + D I ++K GGS+ +++L+ G+ K + G PK K Sbjct: 180 EAVKRVAEKVDDRYKVDEDNIKLEKRQGGSVADTKLVNGIVIDKEVVHPGM---PKRVKN 236 Query: 731 PKISLLNIELELKSEKENAEIRVSSVEEYQRIVDAEWQILYDKLEAIYKSGAKIVLSKLP 910 KI++L LE+K + +AEIR++ ++ + ++ E ++L + ++ + ++GA +V + Sbjct: 237 AKIAVLKAALEVKETETDAEIRITDPDQLMKFIEQEEKMLKEMVDRLAEAGANVVFCQKG 296 Query: 911 IGDVATQYFADRDMFCAGRVTEEDLKRTMAACGGAIQTTVKDITDDTLGKCESFEEVQIG 1090 I D+A Y A + RV + D+++ ACG I T +++IT LG+ E EE ++G Sbjct: 297 IDDLAQYYLAKAGILAVRRVKQSDIEKIAKACGAKIITDLREITSADLGEAELVEERKVG 356 Query: 1091 GERFNIFVGCPEAKTCTIILRGGSEQFMEETERSLHDAIMIVRRAMKNDAVVAGGGAIDM 1270 E+ GC K TI++RGGSE ++E ERSL DAI +V+ A+++ VVAGGGA ++ Sbjct: 357 DEKMVFIEGCKNPKAVTILIRGGSEHVVDEVERSLQDAIKVVKTALESGKVVAGGGAPEI 416 Query: 1271 EISKYLRDYSRNIHGRDQQFIGAMARAFEVIPRQLCENAGFDATNILNKLRQKHAQGKIW 1450 E++ +RD++ + GR+Q A A A EVIPR L EN+G D +IL +LR+ H +GK Sbjct: 417 EVALKIRDWAPTLGGREQLAAEAFASALEVIPRALAENSGLDPIDILVELRKAHEEGKTT 476 Query: 1451 CGVDIMNEDIADNFEACVWEPALVKSNAITAATEAACMVLTLDETIRN---AKSKTPE 1615 GVD+ + ++A E V EP VK+ AIT+ATE A M+L +D+ I K K PE Sbjct: 477 YGVDVFSGEVACMKERGVLEPLKVKTQAITSATEVAIMILRIDDVIAAKGLEKEKGPE 534
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 176,741,640
Number of Sequences: 369166
Number of extensions: 3378862
Number of successful extensions: 11191
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9344
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10239
length of database: 68,354,980
effective HSP length: 116
effective length of database: 46,925,720
effective search space used: 21351202600
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail