DrC_01063
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
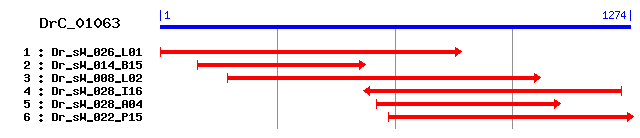
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01063 (1274 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P04634|LIPG_RAT Gastric triacylglycerol lipase precursor... 313 9e-85 sp|P07098|LIPG_HUMAN Gastric triacylglycerol lipase precurs... 310 4e-84 sp|P80035|LIPG_CANFA Gastric triacylglycerol lipase precurs... 310 4e-84 sp|Q9CPP7|LIPG_MOUSE Gastric triacylglycerol lipase precurs... 307 4e-83 sp|Q29458|LIPG_BOVIN Gastric triacylglycerol lipase precurs... 300 5e-81 sp|P38571|LICH_HUMAN Lysosomal acid lipase/cholesteryl este... 296 1e-79 sp|Q9Z0M5|LICH_MOUSE Lysosomal acid lipase/cholesteryl este... 286 9e-77 sp|Q64194|LICH_RAT Lysosomal acid lipase/cholesteryl ester ... 277 5e-74 sp|O46108|LIP3_DROME Lipase 3 precursor (DmLip3) 224 3e-58 sp|O46107|LIP1_DROME Lipase 1 precursor (DmLip1) 222 2e-57
>sp|P04634|LIPG_RAT Gastric triacylglycerol lipase precursor (Gastric lipase) (GL) (Lingual lipase) Length = 395 Score = 313 bits (801), Expect = 9e-85 Identities = 162/364 (44%), Positives = 230/364 (63%), Gaps = 1/364 (0%) Frame = +1 Query: 19 DPEVKMNISEIIRYNGYPCEDHRVVTKDGYILSMQRIPFGSKTRERQTKMTQSRPVVLLQ 198 +PE MNIS++I Y GYPC+++ VVT+DGYIL + RIP G E K RPVV LQ Sbjct: 27 NPEANMNISQMITYWGYPCQEYEVVTEDGYILGVYRIPHGKNNSENIGK----RPVVYLQ 82 Query: 199 HGLLDSAHTWAINSPEQSLAFILADHGFDVWLANSRGNTYSNKHVSLNERDPKFWEFSWD 378 HGL+ SA W N P SLAF+LAD G+DVWL NSRGNT+S K+V + +FW FS+D Sbjct: 83 HGLIASATNWIANLPNNSLAFMLADAGYDVWLGNSRGNTWSRKNVYYSPDSVEFWAFSFD 142 Query: 379 EMASFDLPACVDYALKISGNATKLTYIGHSQGTEIAFAQFSQDVLLRKKISSYVALAPAV 558 EMA +DLPA +++ ++ +G K+ Y+GHSQGT I F FS + L KKI ++ ALAP Sbjct: 143 EMAKYDLPATINFIVQKTGQ-EKIHYVGHSQGTTIGFIAFSTNPTLAKKIKTFYALAPVA 201 Query: 559 FLGNVEAGLRILARFREDIETLLNLFGHGRVMSNSEIFKFLASTICGKEGLPKFCSEVIF 738 + ++ L+ ++ + L +FG + ++ FL + +C +E L CS +F Sbjct: 202 TVKYTQSPLKKISFIPTFLFKL--MFGKKMFLPHTYFDDFLGTEVCSREVLDLLCSNTLF 259 Query: 739 LLAGHDIKNLNESRLPVYISHTPAGTSVMNIVHYAQSVDNGNFQMYDFGIEG-NIKRYGT 915 + G D KNLN SR VY+ H PAGTSV + +H+AQ V +G FQ +++G N+ Y Sbjct: 260 IFCGFDKKNLNVSRFDVYLGHNPAGTSVQDFLHWAQLVRSGKFQAFNWGSPSQNMLHYNQ 319 Query: 916 PNPPQFDVKPVGIPTAIFSGGNDTLADPEDVSRLLGLVGGDVKFQKFISYYNHMDFVWGE 1095 PP++DV + +P A+++GGND LADP+DV+ LL + ++ F K I YNH+DF+W Sbjct: 320 KTPPEYDVSAMTVPVAVWNGGNDILADPQDVAMLLPKL-SNLLFHKEILAYNHLDFIWAM 378 Query: 1096 NAWQ 1107 +A Q Sbjct: 379 DAPQ 382
>sp|P07098|LIPG_HUMAN Gastric triacylglycerol lipase precursor (Gastric lipase) (GL) Length = 398 Score = 310 bits (795), Expect = 4e-84 Identities = 165/364 (45%), Positives = 231/364 (63%), Gaps = 2/364 (0%) Frame = +1 Query: 22 PEVKMNISEIIRYNGYPCEDHRVVTKDGYILSMQRIPFGSKTRERQTKMTQSRPVVLLQH 201 PEV MNIS++I Y GYP E++ VVT+DGYIL + RIP+G K T RPVV LQH Sbjct: 29 PEVTMNISQMITYWGYPNEEYEVVTEDGYILEVNRIPYGKKNSGN----TGQRPVVFLQH 84 Query: 202 GLLDSAHTWAINSPEQSLAFILADHGFDVWLANSRGNTYSNKHVSLNERDPKFWEFSWDE 381 GLL SA W N P SLAFILAD G+DVWL NSRGNT++ +++ + +FW FS+DE Sbjct: 85 GLLASATNWISNLPNNSLAFILADAGYDVWLGNSRGNTWARRNLYYSPDSVEFWAFSFDE 144 Query: 382 MASFDLPACVDYALKISGNATKLTYIGHSQGTEIAFAQFSQDVLLRKKISSYVALAPAVF 561 MA +DLPA +D+ +K +G +L Y+GHSQGT I F FS + L K+I ++ ALAP Sbjct: 145 MAKYDLPATIDFIVKKTGQ-KQLHYVGHSQGTTIGFIAFSTNPSLAKRIKTFYALAP--- 200 Query: 562 LGNVEAGLRILARFREDIETLLN-LFGHGRVMSNSEIFKFLASTICGKEGLPKFCSEVIF 738 + V+ ++ + R ++L +FG ++ +FLA+ +C +E L CS +F Sbjct: 201 VATVKYTKSLINKLRFVPQSLFKFIFGDKIFYPHNFFDQFLATEVCSREMLNLLCSNALF 260 Query: 739 LLAGHDIKNLNESRLPVYISHTPAGTSVMNIVHYAQSVDNGNFQMYDFGIE-GNIKRYGT 915 ++ G D KN N SRL VY+SH PAGTSV N+ H+ Q+V +G FQ YD+G N Y Sbjct: 261 IICGFDSKNFNTSRLDVYLSHNPAGTSVQNMFHWTQAVKSGKFQAYDWGSPVQNRMHYDQ 320 Query: 916 PNPPQFDVKPVGIPTAIFSGGNDTLADPEDVSRLLGLVGGDVKFQKFISYYNHMDFVWGE 1095 PP ++V + +P A+++GG D LADP+DV LL + ++ + K I +YNH+DF+W Sbjct: 321 SQPPYYNVTAMNVPIAVWNGGKDLLADPQDVGLLLPKL-PNLIYHKEIPFYNHLDFIWAM 379 Query: 1096 NAWQ 1107 +A Q Sbjct: 380 DAPQ 383
>sp|P80035|LIPG_CANFA Gastric triacylglycerol lipase precursor (Gastric lipase) (GL) Length = 398 Score = 310 bits (795), Expect = 4e-84 Identities = 165/364 (45%), Positives = 229/364 (62%), Gaps = 1/364 (0%) Frame = +1 Query: 19 DPEVKMNISEIIRYNGYPCEDHRVVTKDGYILSMQRIPFGSKTRERQTKMTQSRPVVLLQ 198 +PEV MNIS++I Y GYP E++ VVT+DGYIL + RIP+G K E + RPV LQ Sbjct: 28 NPEVTMNISQMITYWGYPAEEYEVVTEDGYILGIDRIPYGRKNSENIGR----RPVAFLQ 83 Query: 199 HGLLDSAHTWAINSPEQSLAFILADHGFDVWLANSRGNTYSNKHVSLNERDPKFWEFSWD 378 HGLL SA W N P SLAFILAD G+DVWL NSRGNT++ +++ + +FW FS+D Sbjct: 84 HGLLASATNWISNLPNNSLAFILADAGYDVWLGNSRGNTWARRNLYYSPDSVEFWAFSFD 143 Query: 379 EMASFDLPACVDYALKISGNATKLTYIGHSQGTEIAFAQFSQDVLLRKKISSYVALAPAV 558 EMA +DLPA +D+ LK +G KL Y+GHSQGT I F FS + L K+I ++ ALAP Sbjct: 144 EMAKYDLPATIDFILKKTGQ-DKLHYVGHSQGTTIGFIAFSTNPKLAKRIKTFYALAPVA 202 Query: 559 FLGNVEAGLRILARFREDIETLLNLFGHGRVMSNSEIFKFLASTICGKEGLPKFCSEVIF 738 + E L L + L +FG+ + +FLA+ +C +E + CS +F Sbjct: 203 TVKYTETLLNKLMLVPSFLFKL--IFGNKIFYPHHFFDQFLATEVCSRETVDLLCSNALF 260 Query: 739 LLAGHDIKNLNESRLPVYISHTPAGTSVMNIVHYAQSVDNGNFQMYDFGIE-GNIKRYGT 915 ++ G D NLN SRL VY+SH PAGTSV N++H++Q+V +G FQ +D+G N+ Y Sbjct: 261 IICGFDTMNLNMSRLDVYLSHNPAGTSVQNVLHWSQAVKSGKFQAFDWGSPVQNMMHYHQ 320 Query: 916 PNPPQFDVKPVGIPTAIFSGGNDTLADPEDVSRLLGLVGGDVKFQKFISYYNHMDFVWGE 1095 PP +++ + +P A+++GGND LADP DV LL + + +K I YNH+DF+W Sbjct: 321 SMPPYYNLTDMHVPIAVWNGGNDLLADPHDVDLLLSKLPNLIYHRK-IPPYNHLDFIWAM 379 Query: 1096 NAWQ 1107 +A Q Sbjct: 380 DAPQ 383
>sp|Q9CPP7|LIPG_MOUSE Gastric triacylglycerol lipase precursor (Gastric lipase) (GL) Length = 395 Score = 307 bits (787), Expect = 4e-83 Identities = 160/364 (43%), Positives = 228/364 (62%), Gaps = 1/364 (0%) Frame = +1 Query: 19 DPEVKMNISEIIRYNGYPCEDHRVVTKDGYILSMQRIPFGSKTRERQTKMTQSRPVVLLQ 198 +PE MN+S++I Y GYP E++ VVT+DGYIL + RIP+G K E K RPV LQ Sbjct: 27 NPEANMNVSQMITYWGYPSEEYEVVTEDGYILGVYRIPYGKKNSENIGK----RPVAYLQ 82 Query: 199 HGLLDSAHTWAINSPEQSLAFILADHGFDVWLANSRGNTYSNKHVSLNERDPKFWEFSWD 378 HGL+ SA W N P SLAFILAD G+DVWL NSRGNT+S K+V + +FW FS+D Sbjct: 83 HGLIASATNWITNLPNNSLAFILADAGYDVWLGNSRGNTWSRKNVYYSPDSVEFWAFSFD 142 Query: 379 EMASFDLPACVDYALKISGNATKLTYIGHSQGTEIAFAQFSQDVLLRKKISSYVALAPAV 558 EMA +DLPA +D+ ++ +G K+ Y+GHSQGT I F FS + L KKI + ALAP Sbjct: 143 EMAKYDLPATIDFIVQKTGQ-EKIHYVGHSQGTTIGFIAFSTNPALAKKIKRFYALAPVA 201 Query: 559 FLGNVEAGLRILARFREDIETLLNLFGHGRVMSNSEIFKFLASTICGKEGLPKFCSEVIF 738 + E+ + ++ + + L +FG+ M ++ + +FL + +C +E L CS +F Sbjct: 202 TVKYTESPFKKISLIPKFL--LKVIFGNKMFMPHNYLDQFLGTEVCSRELLDLLCSNALF 259 Query: 739 LLAGHDIKNLNESRLPVYISHTPAGTSVMNIVHYAQSVDNGNFQMYDFGIE-GNIKRYGT 915 + G D KNLN SR VY+ H PAGTS ++ H+AQ +G Q Y++G N+ Y Sbjct: 260 IFCGFDKKNLNVSRFDVYLGHNPAGTSTQDLFHWAQLAKSGKLQAYNWGSPLQNMLHYNQ 319 Query: 916 PNPPQFDVKPVGIPTAIFSGGNDTLADPEDVSRLLGLVGGDVKFQKFISYYNHMDFVWGE 1095 PP +DV + +P A+++GG+D LADP+DV+ LL + ++ + K I YNH+DF+W Sbjct: 320 KTPPYYDVSAMTVPIAVWNGGHDILADPQDVAMLLPKL-PNLLYHKEILPYNHLDFIWAM 378 Query: 1096 NAWQ 1107 +A Q Sbjct: 379 DAPQ 382
>sp|Q29458|LIPG_BOVIN Gastric triacylglycerol lipase precursor (Gastric lipase) (GL) (Pregastric esterase) (PGE) Length = 397 Score = 300 bits (769), Expect = 5e-81 Identities = 159/376 (42%), Positives = 223/376 (59%), Gaps = 1/376 (0%) Frame = +1 Query: 19 DPEVKMNISEIIRYNGYPCEDHRVVTKDGYILSMQRIPFGSKTRERQTKMTQSRPVVLLQ 198 +PE MN+S++I Y GYP E H+V+T DGYIL + RIP G + RPVV LQ Sbjct: 27 NPEASMNVSQMISYWGYPSEMHKVITADGYILQVYRIPHGKNNANHLGQ----RPVVFLQ 82 Query: 199 HGLLDSAHTWAINSPEQSLAFILADHGFDVWLANSRGNTYSNKHVSLNERDPKFWEFSWD 378 HGLL SA W N P+ SL F+LAD G+DVWL NSRGNT++ +H+ + P+FW FS+D Sbjct: 83 HGLLGSATNWISNLPKNSLGFLLADAGYDVWLGNSRGNTWAQEHLYYSPDSPEFWAFSFD 142 Query: 379 EMASFDLPACVDYALKISGNATKLTYIGHSQGTEIAFAQFSQDVLLRKKISSYVALAPAV 558 EMA +DLP+ +D+ L+ +G KL Y+GHSQGT I F FS L +KI + ALAP Sbjct: 143 EMAEYDLPSTIDFILRRTGQ-KKLHYVGHSQGTTIGFIAFSTSPTLAEKIKVFYALAPVA 201 Query: 559 FLGNVEAGLRILARFREDIETLLNLFGHGRVMSNSEIFKFLASTICGKEGLPKFCSEVIF 738 + ++ LA + + +FG ++ + +FL +C +E L C +F Sbjct: 202 TVKYTKSLFNKLALIPHFLFKI--IFGDKMFYPHTFLEQFLGVEMCSRETLDVLCKNALF 259 Query: 739 LLAGHDIKNLNESRLPVYISHTPAGTSVMNIVHYAQSVDNGNFQMYDFGIE-GNIKRYGT 915 + G D KN N SRL VYI+H PAGTSV N +H+ Q+V +G FQ +D+G N+ Y Sbjct: 260 AITGVDNKNFNMSRLDVYIAHNPAGTSVQNTLHWRQAVKSGKFQAFDWGAPYQNLMHYHQ 319 Query: 916 PNPPQFDVKPVGIPTAIFSGGNDTLADPEDVSRLLGLVGGDVKFQKFISYYNHMDFVWGE 1095 P PP +++ + +P A++S ND LADP+DV LL + ++ + K I YNH+DF+W Sbjct: 320 PTPPIYNLTAMNVPIAVWSADNDLLADPQDVDFLLSKL-SNLIYHKEIPNYNHLDFIWAM 378 Query: 1096 NAWQNYLPRCVEFHQE 1143 +A Q V E Sbjct: 379 DAPQEVYNEIVSLMAE 394
>sp|P38571|LICH_HUMAN Lysosomal acid lipase/cholesteryl ester hydrolase precursor (LAL) (Acid cholesteryl ester hydrolase) (Sterol esterase) (Lipase A) (Cholesteryl esterase) Length = 399 Score = 296 bits (757), Expect = 1e-79 Identities = 159/371 (42%), Positives = 228/371 (61%), Gaps = 2/371 (0%) Frame = +1 Query: 7 VTSVDPEVKMNISEIIRYNGYPCEDHRVVTKDGYILSMQRIPFGSKTRERQTKMTQSRPV 186 +T+VDPE MN+SEII Y G+P E++ V T+DGYIL + RIP G K + +PV Sbjct: 26 LTAVDPETNMNVSEIISYWGFPSEEYLVETEDGYILCLNRIPHGRKNHSDKGP----KPV 81 Query: 187 VLLQHGLLDSAHTWAINSPEQSLAFILADHGFDVWLANSRGNTYSNKHVSLNERDPKFWE 366 V LQHGLL + W N SL FILAD GFDVW+ NSRGNT+S KH +L+ +FW Sbjct: 82 VFLQHGLLADSSNWVTNLANSSLGFILADAGFDVWMGNSRGNTWSRKHKTLSVSQDEFWA 141 Query: 367 FSWDEMASFDLPACVDYALKISGNATKLTYIGHSQGTEIAFAQFSQDVLLRKKISSYVAL 546 FS+DEMA +DLPA +++ L +G ++ Y+GHSQGT I F FSQ L K+I + AL Sbjct: 142 FSYDEMAKYDLPASINFILNKTGQ-EQVYYVGHSQGTTIGFIAFSQIPELAKRIKMFFAL 200 Query: 547 APAVFLGNVEAGLRILARFREDIETLLNLFGHGRVMSNSEIFKFLASTICGKEGLPKFCS 726 P + + + L R + + + +LFG + S K+L + +C L + C Sbjct: 201 GPVASVAFCTSPMAKLGRLPDHL--IKDLFGDKEFLPQSAFLKWLGTHVCTHVILKELCG 258 Query: 727 EVIFLLAGHDIKNLNESRLPVYISHTPAGTSVMNIVHYAQSVDNGNFQMYDFGIEG-NIK 903 + FLL G + +NLN SR+ VY +H+PAGTSV N++H++Q+V FQ +D+G N Sbjct: 259 NLCFLLCGFNERNLNMSRVDVYTTHSPAGTSVQNMLHWSQAVKFQKFQAFDWGSSAKNYF 318 Query: 904 RYGTPNPPQFDVKPVGIPTAIFSGGNDTLADPEDVSRLLGLVGGDVKFQKFISYYNHMDF 1083 Y PP ++VK + +PTA++SGG+D LAD DV+ LL + ++ F + I + H+DF Sbjct: 319 HYNQSYPPTYNVKDMLVPTAVWSGGHDWLADVYDVNILLTQI-TNLVFHESIPEWEHLDF 377 Query: 1084 VWGENA-WQNY 1113 +WG +A W+ Y Sbjct: 378 IWGLDAPWRLY 388
>sp|Q9Z0M5|LICH_MOUSE Lysosomal acid lipase/cholesteryl ester hydrolase precursor (LAL) (Acid cholesteryl ester hydrolase) (Sterol esterase) (Lipase A) (Cholesteryl esterase) Length = 397 Score = 286 bits (732), Expect = 9e-77 Identities = 159/371 (42%), Positives = 224/371 (60%), Gaps = 2/371 (0%) Frame = +1 Query: 7 VTSVDPEVKMNISEIIRYNGYPCEDHRVVTKDGYILSMQRIPFGSKTRERQTKMTQSRPV 186 V++VDPEV MN++EII GYP E+H V+T DGYILS+ RIP G K + RPV Sbjct: 24 VSAVDPEVNMNVTEIIMRWGYPGEEHSVLTGDGYILSIHRIPRGWKNHFGKGP----RPV 79 Query: 187 VLLQHGLLDSAHTWAINSPEQSLAFILADHGFDVWLANSRGNTYSNKHVSLNERDPKFWE 366 V LQHGLL + W N SL F+LAD GFDVW+ NSRGNT+S KH +L+ +FW Sbjct: 80 VYLQHGLLADSSNWVTNIDNSSLGFLLADRGFDVWMGNSRGNTWSLKHKTLSVSQDEFWA 139 Query: 367 FSWDEMASFDLPACVDYALKISGNATKLTYIGHSQGTEIAFAQFSQDVLLRKKISSYVAL 546 FS+DEMA +DLPA ++Y L +G ++ Y+GHSQG I F FSQ L KKI ++ L Sbjct: 140 FSFDEMAKYDLPASINYILNKTGQ-EQIYYVGHSQGCTIGFIAFSQMPELAKKIKMFLVL 198 Query: 547 APAVFLGNVEAGLRILARFREDIETLLNLFGHGRVMSNSEIFKFLASTICGKEGLPKFCS 726 AP + L L L R + + L ++FG + + S + K+L+ +C + + C+ Sbjct: 199 APVLSLNFASGPLLQLGRLPDPL--LKDMFGQKQFLPQSAMLKWLSIHVCTHVIMKELCA 256 Query: 727 EVIFLLAGHDIKNLNESRLPVYISHTPAGTSVMNIVHYAQSVDNGNFQMYDFG-IEGNIK 903 V FLL G + KNLN SR+ VY +H PA V N++H+ Q Q +D+G E N Sbjct: 257 NVFFLLCGFNEKNLNMSRVDVYTTHCPAELLVQNMLHWGQVFKYRKLQAFDWGSSEKNYF 316 Query: 904 RYGTPNPPQFDVKPVGIPTAIFSGGNDTLADPEDVSRLLGLVGGDVKFQKFISYYNHMDF 1083 Y PP +++K + +PTA++SGG D LAD D++ LL + + + K I ++H+DF Sbjct: 317 HYNQSFPPSYNIKNMRLPTALWSGGRDWLADINDITILLTQI-PKLVYHKNIPEWDHLDF 375 Query: 1084 VWGENA-WQNY 1113 +WG +A W+ Y Sbjct: 376 IWGLDAPWKLY 386
>sp|Q64194|LICH_RAT Lysosomal acid lipase/cholesteryl ester hydrolase precursor (LAL) (Acid cholesteryl ester hydrolase) (Sterol esterase) (Lipase A) (Cholesteryl esterase) Length = 397 Score = 277 bits (708), Expect = 5e-74 Identities = 156/373 (41%), Positives = 224/373 (60%), Gaps = 4/373 (1%) Frame = +1 Query: 7 VTSVDPEVKMNISEIIRYNGYPCEDHRVVTKDGYILSMQRIPFGSKTRERQTKMTQSRPV 186 +++VDPE MN++EII + GYP +H V T DGYIL + RIP G K + + +PV Sbjct: 24 ISAVDPEANMNVTEIIMHWGYP--EHSVQTGDGYILGVHRIPHGRKNQFDKGP----KPV 77 Query: 187 VLLQ--HGLLDSAHTWAINSPEQSLAFILADHGFDVWLANSRGNTYSNKHVSLNERDPKF 360 V LQ HG L + W N SL FILAD GFDVW+ NSRGNT+S KH +L+ ++ Sbjct: 78 VYLQWRHGFLADSSNWVTNIDNNSLGFILADAGFDVWMGNSRGNTWSRKHKTLSVSQDEY 137 Query: 361 WEFSWDEMASFDLPACVDYALKISGNATKLTYIGHSQGTEIAFAQFSQDVLLRKKISSYV 540 W FS+DEMA +DLPA ++Y L +G +L +GHSQG I F FSQ L KK+ + Sbjct: 138 WAFSFDEMAKYDLPASINYILNKTGQ-EQLYNVGHSQGCTIGFIAFSQMPELAKKVKMFF 196 Query: 541 ALAPAVFLGNVEAGLRILARFREDIETLLNLFGHGRVMSNSEIFKFLASTICGKEGLPKF 720 ALAP + L + L R + + L +LFG + + S + K+L++ IC + + Sbjct: 197 ALAPVLSLNFASGPMVKLGRLPDLL--LEDLFGQKQFLPQSAMVKWLSTHICTHVIMKEL 254 Query: 721 CSEVIFLLAGHDIKNLNESRLPVYISHTPAGTSVMNIVHYAQSVDNGNFQMYDFG-IEGN 897 C+ + FL+ G + KNLN SR+ VY +H PAGTSV N+VH+ Q V Q +D+G + N Sbjct: 255 CANIFFLICGFNEKNLNMSRVDVYTTHCPAGTSVQNMVHWTQVVKYHKLQAFDWGSSDKN 314 Query: 898 IKRYGTPNPPQFDVKPVGIPTAIFSGGNDTLADPEDVSRLLGLVGGDVKFQKFISYYNHM 1077 Y PP + +K + +PTA++SGG D LAD D++ LL + + + K I ++H+ Sbjct: 315 YFHYNQSYPPLYSIKDMQLPTALWSGGKDWLADTSDINILLTEI-PTLVYHKNIPEWDHL 373 Query: 1078 DFVWGENA-WQNY 1113 DF+WG +A W+ Y Sbjct: 374 DFIWGLDAPWRLY 386
>sp|O46108|LIP3_DROME Lipase 3 precursor (DmLip3) Length = 394 Score = 224 bits (572), Expect = 3e-58 Identities = 129/359 (35%), Positives = 193/359 (53%), Gaps = 7/359 (1%) Frame = +1 Query: 46 EIIRYNGYPCEDHRVVTKDGYILSMQRIPFGSKTRERQTKMTQSRPVVLLQHGLLDSAHT 225 E I +GYP E H VVT D YIL+M RIP+ KT E + +RPV L HG+L S+ Sbjct: 30 ERIEDDGYPMERHEVVTSDNYILTMHRIPYSPKTGE-----SSNRPVAFLMHGMLSSSSD 84 Query: 226 WAINSPEQSLAFILADHGFDVWLANSRGNTYSNKHVSLNERDPKFWEFSWDEMASFDLPA 405 W + PE+SLA++LAD G+DVW+ N+RGNTYS H FW FSW+E+ +D+PA Sbjct: 85 WVLMGPERSLAYMLADAGYDVWMGNARGNTYSKAHKYWPTYWQIFWNFSWNEIGMYDVPA 144 Query: 406 CVDYALKISGNATKLTYIGHSQGTEIAFAQFSQDVLLRKKISSYVALAPAVFLGNVEAGL 585 +DY L +G ++ Y+GHSQGT + S+ KI S L PA ++GN+++ L Sbjct: 145 MIDYVLAKTGQ-QQVQYVGHSQGTTVYLVMVSERPEYNDKIKSAHLLGPAAYMGNMKSPL 203 Query: 586 -RILARFREDIETLLNLFGHGRVMSNSEIFKFLASTIC-GKEGLPKFCSEVIFLLAGHDI 759 R A ++ + G M +++ + L +C C+ IFL+ G+D Sbjct: 204 TRAFAPILGQPNAIVEVCGSMEFMPSNKFKQDLGIEMCQATSPYADMCANEIFLIGGYDT 263 Query: 760 KNLNESRLPVYISHTPAGTSVMNIVHYAQSVDNGNFQMYDFGIEGNIKRYGTPNPPQFDV 939 + L+ L + +PAG SV +H+ Q ++G F+ +D+ N YG+ PP + + Sbjct: 264 EQLDYELLEHIKATSPAGASVNQNLHFCQEYNSGKFRKFDYTALRNPYEYGSYFPPDYKL 323 Query: 940 KPVGIPTAIFSGGNDTLADPEDVSRL---LGLVGGD--VKFQKFISYYNHMDFVWGENA 1101 K P ++ G ND + D DV +L L + D V F+K + H+DF+WG A Sbjct: 324 KNAKAPVLLYYGANDWMCDVSDVRKLRDELPNMALDYLVPFEK----WAHLDFIWGTEA 378
>sp|O46107|LIP1_DROME Lipase 1 precursor (DmLip1) Length = 439 Score = 222 bits (565), Expect = 2e-57 Identities = 133/364 (36%), Positives = 189/364 (51%), Gaps = 5/364 (1%) Frame = +1 Query: 13 SVDPEVKMNISEIIRYNGYPCEDHRVVTKDGYILSMQRIPFGSKTRERQTKMTQSRPVVL 192 ++ + +++ ++I GY E H V T+DGYIL+M RI Q P L Sbjct: 59 NIKQDSTLSVDKLIAKYGYESEVHHVTTEDGYILTMHRI------------RKQGAPPFL 106 Query: 193 LQHGLLDSAHTWAINSPEQSLAFILADHGFDVWLANSRGNTYSNKHVSLNERDPKFWEFS 372 LQHGL+DS+ + + P SLA++LADH +DVWL N+RGN YS H +L+ + KFW+FS Sbjct: 107 LQHGLVDSSAGFVVMGPNVSLAYLLADHNYDVWLGNARGNRYSRNHTTLDPDESKFWDFS 166 Query: 373 WDEMASFDLPACVDYALKISGNATKLTYIGHSQGTEIAFAQFSQDVLLRKKISSYVALAP 552 W E+ +DLPA +D+ LK++G KL Y GHSQG F S K+ S ALAP Sbjct: 167 WHEIGMYDLPAMIDHVLKVTG-FPKLHYAGHSQGCTSFFVMCSMRPAYNDKVVSMQALAP 225 Query: 553 AVFLGNVEAGLRILARFREDIETLLN-LFGHG-RVMSNSEIFKFLASTICGKEGLPKFCS 726 AV+ E I A I N L G R M N E F+FL E + C Sbjct: 226 AVYAKETEDHPYIRA-----ISLYFNSLVGSSIREMFNGE-FRFLCRM---TEETERLCI 276 Query: 727 EVIFLLAGHDIKNLNESRLPVYISHTPAGTSVMNIVHYAQSVDNGNFQMYDFGIEGNIKR 906 E +F + G + N PV + H PAG + + H+ Q + +G F Y + N++ Sbjct: 277 EAVFGIVGRNWNEFNRKMFPVILGHYPAGVAAKQVKHFIQIIKSGRFAPYSYSSNKNMQL 336 Query: 907 YGTPNPPQFDVKPVGIPTAIFSGGNDTLADPEDVSRL---LGLVGGDVKFQKFISYYNHM 1077 Y PP++++ V +PT ++ ND L P+DV + LG V G K+ +NHM Sbjct: 337 YRDHLPPRYNLSLVTVPTFVYYSTNDLLCHPKDVESMCDDLGNVTG--KYLVPQKEFNHM 394 Query: 1078 DFVW 1089 DF+W Sbjct: 395 DFLW 398
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 165,772,008
Number of Sequences: 369166
Number of extensions: 3837122
Number of successful extensions: 9783
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9130
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9728
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 14766256675
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail