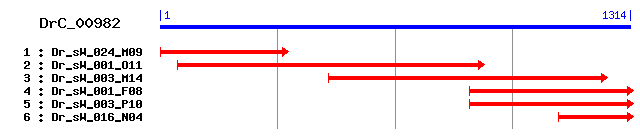
DrC_00982
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00982 (1314 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q8BKZ9|ODPX_MOUSE Pyruvate dehydrogenase protein X compo... 275 3e-73 sp|O00330|ODPX_HUMAN Pyruvate dehydrogenase protein X compo... 271 2e-72 sp|O66119|ODP2_ZYMMO Dihydrolipoyllysine-residue acetyltran... 246 1e-64 sp|P08461|ODP2_RAT Dihydrolipoyllysine-residue acetyltransf... 245 2e-64 sp|O59816|ODP2_SCHPO Dihydrolipoyllysine-residue acetyltran... 239 1e-62 sp|P10515|ODP2_HUMAN Dihydrolipoyllysine-residue acetyltran... 238 2e-62 sp|Q9ZD20|ODP2_RICPR Dihydrolipoyllysine-residue acetyltran... 236 1e-61 sp|P36413|ODP2_DICDI Dihydrolipoyllysine-residue acetyltran... 234 5e-61 sp|Q92HK7|ODP2_RICCN Dihydrolipoyllysine-residue acetyltran... 229 1e-59 sp|Q9R9N3|ODP2_RHIME Dihydrolipoyllysine-residue acetyltran... 225 2e-58
>sp|Q8BKZ9|ODPX_MOUSE Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X) Length = 501 Score = 275 bits (702), Expect = 3e-73 Identities = 158/441 (35%), Positives = 251/441 (56%), Gaps = 32/441 (7%) Frame = +1 Query: 13 IKMPSLSPTMSEGGIVEWYKKEGDKISAGDLICDVQTDKAVVGMEADEEGILAKILKAKD 192 + MPSLSPTM +G IV+W +KEG+ +SAGD +C+++TDKAVV ++A+++GILAKI+ + Sbjct: 59 VLMPSLSPTMEQGNIVKWLRKEGEAVSAGDSLCEIETDKAVVTLDANDDGILAKIVVEEG 118 Query: 193 SQGIQVGEAIALLAGEDEDWKTVEIPAGGATVGVXXXXXXXXXXXXXXXXXXXXRKSNKA 372 ++ IQ+G IAL+ E EDWK VEIP + RK +K Sbjct: 119 AKNIQLGSLIALMVEEGEDWKQVEIPKDVSAPPPVSKPPAPTQPSPQPQIPCPARKEHKG 178 Query: 373 TS--KLGPAVRRLLDTYGINENQVDHSGPHDVLTKSDVLQ--------HIDRSRLSPVCP 522 T+ +L PA R +L+ + ++ +Q +GP + TK D L+ I SR + P Sbjct: 179 TARFRLSPAARNILEKHSLDASQGTATGPRGIFTKEDALKLVELKQMGKITESRPASAPP 238 Query: 523 PTTRPAAQPEXXXXXXH--------------------VDVPLTNMRQTIARRMAASKSSI 642 P+ + P+ + ++P +N+R+ IA+R+ SKS++ Sbjct: 239 PSLSASVPPQATAGPSYPRPMTPPVSIPGQPNAAGTFTEIPASNIRRVIAKRLTESKSTV 298 Query: 643 PHRYSTAEASXXXXXXXXXXXXDQRGLVLSVNDFIVKAASLALRLVPEINNGLNADGTIK 822 PH Y+TA+ + + +SVNDFI++AA++ L+ +P +N + +G + Sbjct: 299 PHAYATADCDLGAVLKVRRDLV-KDDIKVSVNDFIIRAAAVTLKQMPGVNVTWDGEGPKQ 357 Query: 823 INHNIDISVAVASESGLITPIVFGADKLPLAAISSTIHELAEKARNNKLQPQEFMGGTFT 1002 + ++DISVAVA++ GLITPI+ A + I+ ++ L++KAR+ KL P+E+ GG+F+ Sbjct: 358 L-PSVDISVAVATDKGLITPIIKDAAAKGIQEIADSVKVLSKKARDGKLMPEEYQGGSFS 416 Query: 1003 ISNLGMFGIKEFTAIINSPQAAILAVGAGIP--HLAHDPANPSKPKSVTKMRFTLCADAR 1176 ISNLGMFGI EF A+IN PQA ILAVG P L D + + + T+ +D+R Sbjct: 417 ISNLGMFGIDEFAAVINPPQACILAVGRFRPVLKLTEDEEGNPQLQQHQLITVTMSSDSR 476 Query: 1177 IIDETSASEFVSRVKSYLENP 1239 ++D+ A+ F+ K+ LENP Sbjct: 477 VVDDELATRFLETFKANLENP 497
>sp|O00330|ODPX_HUMAN Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X) (E3-binding protein) (E3BP) (proX) Length = 501 Score = 271 bits (694), Expect = 2e-72 Identities = 159/441 (36%), Positives = 244/441 (55%), Gaps = 32/441 (7%) Frame = +1 Query: 13 IKMPSLSPTMSEGGIVEWYKKEGDKISAGDLICDVQTDKAVVGMEADEEGILAKILKAKD 192 I MPSLSPTM EG IV+W KKEG+ +SAGD +C+++TDKAVV ++A ++GILAKI+ + Sbjct: 59 ILMPSLSPTMEEGNIVKWLKKEGEAVSAGDALCEIETDKAVVTLDASDDGILAKIVVEEG 118 Query: 193 SQGIQVGEAIALLAGEDEDWKTVEIPAGGATVGVXXXXXXXXXXXXXXXXXXXXRKSNKA 372 S+ I++G I L+ E EDWK VEIP ++ Sbjct: 119 SKNIRLGSLIGLIVEEGEDWKHVEIPKDVGPPPPVSKPSEPRPSPEPQISIPVKKEHIPG 178 Query: 373 TSK--LGPAVRRLLDTYGINENQVDHSGPHDVLTKSDVLQHIDRSRLSPVCP--PTTRPA 540 T + L PA R +L+ + ++ +Q +GP + TK D L+ + + + PT P Sbjct: 179 TLRFRLSPAARNILEKHSLDASQGTATGPRGIFTKEDALKLVQLKQTGKITESRPTPAPT 238 Query: 541 AQP--------------------------EXXXXXXHVDVPLTNMRQTIARRMAASKSSI 642 A P + ++P +N+R+ IA+R+ SKS++ Sbjct: 239 ATPTAPSPLQATAGPSYPRPVIPPVSTPGQPNAVGTFTEIPASNIRRVIAKRLTESKSTV 298 Query: 643 PHRYSTAEASXXXXXXXXXXXXDQRGLVLSVNDFIVKAASLALRLVPEINNGLNADGTIK 822 PH Y+TA+ + +SVNDFI+KAA++ L+ +P++N + +G + Sbjct: 299 PHAYATADCDLGAVLKVRQDLVKD-DIKVSVNDFIIKAAAVTLKQMPDVNVSWDGEGPKQ 357 Query: 823 INHNIDISVAVASESGLITPIVFGADKLPLAAISSTIHELAEKARNNKLQPQEFMGGTFT 1002 + IDISVAVA++ GL+TPI+ A + I+ ++ L++KAR+ KL P+E+ GG+F+ Sbjct: 358 LPF-IDISVAVATDKGLLTPIIKDAAAKGIQEIADSVKALSKKARDGKLLPEEYQGGSFS 416 Query: 1003 ISNLGMFGIKEFTAIINSPQAAILAVGAGIP--HLAHDPANPSKPKSVTKMRFTLCADAR 1176 ISNLGMFGI EFTA+IN PQA ILAVG P L D +K + + T+ +D+R Sbjct: 417 ISNLGMFGIDEFTAVINPPQACILAVGRFRPVLKLTEDEEGNAKLQQRQLITVTMSSDSR 476 Query: 1177 IIDETSASEFVSRVKSYLENP 1239 ++D+ A+ F+ K+ LENP Sbjct: 477 VVDDELATRFLKSFKANLENP 497
>sp|O66119|ODP2_ZYMMO Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex (E2) (Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex) Length = 440 Score = 246 bits (627), Expect = 1e-64 Identities = 153/441 (34%), Positives = 228/441 (51%), Gaps = 27/441 (6%) Frame = +1 Query: 13 IKMPSLSPTMSEGGIVEWYKKEGDKISAGDLICDVQTDKAVVGMEADEEGILAKILKAKD 192 +KMP+LSPTM+EG + +W KEGD + AGD++ +++TDKA++ E + GI+AKIL + Sbjct: 5 VKMPALSPTMTEGTLAKWLVKEGDAVKAGDILAEIETDKAIMEFETVDAGIIAKILVPEG 64 Query: 193 SQGIQVGEAIALLAGEDEDWKTVEIPAGGATVGVXXXXXXXXXXXXXXXXXXXXRKSNKA 372 S+ I VG+ IA++A ED V A +KA Sbjct: 65 SENIAVGQVIAVMAEAGEDVSQVAASASSQISEPSEKADVAQKETADSETISIDASLDKA 124 Query: 373 TSKLG----------------------PAVRRLLDTYGINENQVDHSGPHDVLTKSDVLQ 486 S G P +RL ++ QV+ SGPH + K+D+ Sbjct: 125 ISNAGYGNKTENMTASYQEKAGRIKASPLAKRLAKKNHVDLKQVNGSGPHGRIIKADIEA 184 Query: 487 HIDRSRLSPVCPPTTRPAAQPEXXXXXXHVDVPLTNMRQTIARRMAASKSSIPHRYSTAE 666 I + + P + P A + H + L+NMR+ IARR+ SK +IPH Y T + Sbjct: 185 FIAEANQASSNPSVSTPEASGKITHDTPHNSIKLSNMRRVIARRLTESKQNIPHIYLTVD 244 Query: 667 ASXXXXXXXXXXXXDQ---RGLVLSVNDFIVKAASLALRLVPEINNGLNADGTIKINHNI 837 + + + +SVND ++KA +LAL+ P +N + D ++ + Sbjct: 245 VQMDALLKLRSELNESLAVQNIKISVNDMLIKAQALALKATPNVNVAFDGDQMLQFSQ-A 303 Query: 838 DISVAVASESGLITPIVFGADKLPLAAISSTIHELAEKARNNKLQPQEFMGGTFTISNLG 1017 DISVAV+ E GLITPI+ AD L+A+S + EL +AR +LQPQE+ GGT +ISN+G Sbjct: 304 DISVAVSVEGGLITPILKQADTKSLSALSVEMKELIARAREGRLQPQEYQGGTSSISNMG 363 Query: 1018 MFGIKEFTAIINSPQAAILAVGAG--IPHLAHDPANPSKPKSVTKMRFTLCADARIIDET 1191 MFGIK+F A+IN PQA+ILA+G+G P + D + ++T D R+ID Sbjct: 364 MFGIKQFNAVINPPQASILAIGSGERRPWVIDDAITIATVATITG-----SFDHRVIDGA 418 Query: 1192 SASEFVSRVKSYLENPSFLLA 1254 A+ F+S K +E P +LA Sbjct: 419 DAAAFMSAFKHLVEKPLGILA 439
>sp|P08461|ODP2_RAT Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex (E2) (Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex) (PDC-E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) Length = 555 Score = 245 bits (626), Expect = 2e-64 Identities = 155/426 (36%), Positives = 230/426 (53%), Gaps = 13/426 (3%) Frame = +1 Query: 13 IKMPSLSPTMSEGGIVEWYKKEGDKISAGDLICDVQTDKAVVGMEADEEGILAKILKAKD 192 I +P+LSPTM+ G + W KK G+K+S GDL+ +++TDKA +G E EEG LAKIL + Sbjct: 134 IVLPALSPTMTMGTVQRWEKKVGEKLSEGDLLAEIETDKATIGFEVQEEGYLAKILVPEG 193 Query: 193 SQGIQVGEAIALLAGEDED------WKTVEIPA--GGATVGVXXXXXXXXXXXXXXXXXX 348 ++ + +G + ++ + ED ++ E+ + A V Sbjct: 194 TRDVPLGTPLCIIVEKQEDIAAFADYRPTEVTSLKPQAPPPVPPPVAAVPPIPQPLAPTP 253 Query: 349 XXRKSN-KATSKLGPAVRRLLDTYGINENQVDHSGPHDVLTKSDVLQHIDRSRLSPVCPP 525 + K + P ++L GI+ QV +GP + K D+ + ++ +P Sbjct: 254 SAAPAGPKGRVFVSPLAKKLAAEKGIDLTQVKGTGPEGRIIKKDIDSFVP-TKAAPAAAA 312 Query: 526 TTRPAAQPEXXXXXXHVDVPLTNMRQTIARRMAASKSSIPHRYSTAEASXXXXXXXXXXX 705 P + +D+P++N+R+ IA+R+ SK +IPH Y + + + Sbjct: 313 AAPPGPRVAPTPAGVFIDIPISNIRRVIAQRLMQSKQTIPHYYLSVDVNMGEVLLVRKEL 372 Query: 706 X---DQRGLVLSVNDFIVKAASLALRLVPEINNGLNADGTIKINHNIDISVAVASESGLI 876 + +G + SVNDFI+KA++LA VPE N+ D I+ NH +D+SVAV++ +GLI Sbjct: 373 NKMLEGKGKI-SVNDFIIKASALACLKVPEANSSW-MDTVIRQNHVVDVSVAVSTPAGLI 430 Query: 877 TPIVFGADKLPLAAISSTIHELAEKARNNKLQPQEFMGGTFTISNLGMFGIKEFTAIINS 1056 TPIVF A L I+S + LA KAR KLQP EF GGTFTISNLGMFGIK F+AIIN Sbjct: 431 TPIVFNAHIKGLETIASDVVSLASKAREGKLQPHEFQGGTFTISNLGMFGIKNFSAIINP 490 Query: 1057 PQAAILAVGAGIPHLAHDPANPSKPKSVTK-MRFTLCADARIIDETSASEFVSRVKSYLE 1233 PQA ILA+GA L PA+ K V M TL D R++D +++++ K YLE Sbjct: 491 PQACILAIGASEDKLI--PADNEKGFDVASVMSVTLSCDHRVVDGAVGAQWLAEFKKYLE 548 Query: 1234 NPSFLL 1251 P +L Sbjct: 549 KPVTML 554
Score = 78.2 bits (191), Expect = 5e-14
Identities = 38/79 (48%), Positives = 53/79 (67%)
Frame = +1
Query: 13 IKMPSLSPTMSEGGIVEWYKKEGDKISAGDLICDVQTDKAVVGMEADEEGILAKILKAKD 192
+ +PSLSPTM G I W KKEG+KIS GDLI +V+TDKA VG E+ EE +AKIL +
Sbjct: 8 VPLPSLSPTMQAGTIARWEKKEGEKISEGDLIAEVETDKATVGFESLEECYMAKILVPEG 67
Query: 193 SQGIQVGEAIALLAGEDED 249
++ + +G I + + +D
Sbjct: 68 TRDVPIGCIICITVEKPQD 86
>sp|O59816|ODP2_SCHPO Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (E2) (Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex) (PDC-E2) Length = 483 Score = 239 bits (611), Expect = 1e-62 Identities = 155/432 (35%), Positives = 223/432 (51%), Gaps = 21/432 (4%) Frame = +1 Query: 7 TLIKMPSLSPTMSEGGIVEWYKKEGDKISAGDLICDVQTDKAVVGMEADEEGILAKILKA 186 T+I MP+LSPTM+ G I + KK GDKI GD++C+++TDKA + E +EG LAKIL Sbjct: 54 TVINMPALSPTMTTGNIGAFQKKIGDKIEPGDVLCEIETDKAQIDFEQQDEGYLAKILIE 113 Query: 187 KDSQGIQVGEAIALLAGEDEDWKTVE---IPAGGATVGVXXXXXXXXXXXXXXXXXXXXR 357 ++ + VG+ +A+ + D + I A Sbjct: 114 TGTKDVPVGKPLAVTVENEGDVAAMADFTIEDSSAKEPSAKSGEEKSAPSSEKQSKETSS 173 Query: 358 KSNKATSKLG------PAVRRLLDTYGINENQVDHSGPHDVLTKSDVLQHIDRSRLSPVC 519 SN + + G P R+L + ++ +Q+ SGP+ + K D+ PV Sbjct: 174 PSNVSGEERGDRVFASPLARKLAEEKDLDLSQIRGSGPNGRIIKVDI------ENFKPVV 227 Query: 520 PP----------TTRPAAQPEXXXXXXHVDVPLTNMRQTIARRMAASKSSIPHRYSTAEA 669 P TT A+ + + D+PL+NMR+ IA R+A SK+ PH Y T Sbjct: 228 APKPSNEAAAKATTPAASAADAAAPGDYEDLPLSNMRKIIASRLAESKNMNPHYYVTVSV 287 Query: 670 SXXXXXXXXXXXXDQRG--LVLSVNDFIVKAASLALRLVPEINNGLNADGTIKINHNIDI 843 + LSVND ++KA + ALR VPE+N D I+ N+DI Sbjct: 288 NMEKIIRLRAALNAMADGRYKLSVNDLVIKATTAALRQVPEVNAAWMGD-FIRQYKNVDI 346 Query: 844 SVAVASESGLITPIVFGADKLPLAAISSTIHELAEKARNNKLQPQEFMGGTFTISNLGMF 1023 S+AVA+ SGLITP++ L LA IS+ + ++ARNNKL+P+E+ GGTFTISNLGMF Sbjct: 347 SMAVATPSGLITPVIRNTHALGLAEISTLAKDYGQRARNNKLKPEEYQGGTFTISNLGMF 406 Query: 1024 GIKEFTAIINSPQAAILAVGAGIPHLAHDPANPSKPKSVTKMRFTLCADARIIDETSASE 1203 + +FTAIIN PQA ILAVG + + D + K M+ TL +D R++D A+ Sbjct: 407 PVDQFTAIINPPQACILAVGTTVDTVVPDSTSEKGFKVAPIMKCTLSSDHRVVDGAMAAR 466 Query: 1204 FVSRVKSYLENP 1239 F + +K LENP Sbjct: 467 FTTALKKILENP 478
>sp|P10515|ODP2_HUMAN Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S-acetyltransferase component of pyruvate dehydrogenase complex) (PDC-E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit) Length = 614 Score = 238 bits (608), Expect = 2e-62 Identities = 154/433 (35%), Positives = 229/433 (52%), Gaps = 20/433 (4%) Frame = +1 Query: 13 IKMPSLSPTMSEGGIVEWYKKEGDKISAGDLICDVQTDKAVVGMEADEEGILAKILKAKD 192 + +P+LSPTM+ G + W KK G+K+S GDL+ +++TDKA +G E EEG LAKIL + Sbjct: 188 VLLPALSPTMTMGTVQRWEKKVGEKLSEGDLLAEIETDKATIGFEVQEEGYLAKILVPEG 247 Query: 193 SQGIQVGEAIALLAGEDEDWKTV-------------EIPAGGATVGVXXXXXXXXXXXXX 333 ++ + +G + ++ ++ D ++P Sbjct: 248 TRDVPLGTPLCIIVEKEADISAFADYRPTEVTDLKPQVPPPTPPPVAAVPPTPQPLAPTP 307 Query: 334 XXXXXXXRKSNKATSKLGPAVRRLLDTYGINENQVDHSGPHDVLTKSDVLQHIDRSRLSP 513 K + P ++L GI+ QV +GP +TK D+ + S+++P Sbjct: 308 STPCPATPAGPKGRVFVDPLAKKLAVEKGIDLTQVKGTGPDGRITKKDIDSFVP-SKVAP 366 Query: 514 ----VCPPTTRPAAQPEXXXXXXHVDVPLTNMRQTIARRMAASKSSIPHRYSTAEASXXX 681 V PPT P P D+P++N+R+ IA+R+ SK +IPH Y + + + Sbjct: 367 APAAVVPPTG-PGMAP--VPTGVFTDIPISNIRRVIAQRLMQSKQTIPHYYLSIDVNMGE 423 Query: 682 XXXXXXXXXD--QRGLVLSVNDFIVKAASLALRLVPEINNGLNADGTIKINHNIDISVAV 855 + +SVNDFI+KA++LA VPE N+ D I+ NH +D+SVAV Sbjct: 424 VLLVRKELNKILEGRSKISVNDFIIKASALACLKVPEANSSW-MDTVIRQNHVVDVSVAV 482 Query: 856 ASESGLITPIVFGADKLPLAAISSTIHELAEKARNNKLQPQEFMGGTFTISNLGMFGIKE 1035 ++ +GLITPIVF A + I++ + LA KAR KLQP EF GGTFTISNLGMFGIK Sbjct: 483 STPAGLITPIVFNAHIKGVETIANDVVSLATKAREGKLQPHEFQGGTFTISNLGMFGIKN 542 Query: 1036 FTAIINSPQAAILAVGAGIPHLAHDPANPSKPKSV-TKMRFTLCADARIIDETSASEFVS 1212 F+AIIN PQA ILA+GA L PA+ K V + M TL D R++D +++++ Sbjct: 543 FSAIINPPQACILAIGASEDKLV--PADNEKGFDVASMMSVTLSCDHRVVDGAVGAQWLA 600 Query: 1213 RVKSYLENPSFLL 1251 + YLE P +L Sbjct: 601 EFRKYLEKPITML 613
Score = 85.9 bits (211), Expect = 2e-16
Identities = 41/79 (51%), Positives = 55/79 (69%)
Frame = +1
Query: 13 IKMPSLSPTMSEGGIVEWYKKEGDKISAGDLICDVQTDKAVVGMEADEEGILAKILKAKD 192
+ +PSLSPTM G I W KKEGDKI+ GDLI +V+TDKA VG E+ EE +AKIL A+
Sbjct: 61 VPLPSLSPTMQAGTIARWEKKEGDKINEGDLIAEVETDKATVGFESLEECYMAKILVAEG 120
Query: 193 SQGIQVGEAIALLAGEDED 249
++ + +G I + G+ ED
Sbjct: 121 TRDVPIGAIICITVGKPED 139
>sp|Q9ZD20|ODP2_RICPR Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex (E2) (Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex) Length = 408 Score = 236 bits (601), Expect = 1e-61 Identities = 149/420 (35%), Positives = 223/420 (53%), Gaps = 7/420 (1%) Frame = +1 Query: 13 IKMPSLSPTMSEGGIVEWYKKEGDKISAGDLICDVQTDKAVVGMEADEEGILAKILKAKD 192 I MP+LSPTM EG + W KKEGDK++ G++I +++TDKA + +E+ +EGILAKI+ ++ Sbjct: 5 ILMPALSPTMREGNLARWLKKEGDKVNPGEVIAEIETDKATMEVESVDEGILAKIIIPQN 64 Query: 193 SQGIQVGEAIALLAGEDEDWKTVE---IPAGGATVGVXXXXXXXXXXXXXXXXXXXXRKS 363 SQ + V IA+L+ E ED ++ ++ + S Sbjct: 65 SQNVPVNSLIAVLSEEGEDKADIDSFIAQNNSVSLSLKTDATLKKSNDSITNVEGIKHDS 124 Query: 364 NKATSKLGPAVRRLLDTYGINENQVDHSGPHDVLTKSDVLQHIDRSRLSPVCPPTTRPAA 543 NK + P +RL I V SGPH + K D+L + + + + T Sbjct: 125 NKIFAS--PLAKRLAKIGDIRLENVQGSGPHGRIVKQDILSYDSSTSSNKIVYRDTEE-- 180 Query: 544 QPEXXXXXXHVDVPLTNMRQTIARRMAASKSSIPHRYSTAEASXXXXXXXXXXXX----D 711 + VP N+R+ IA+R+ SK ++PH Y + E + + Sbjct: 181 ---------YRSVPNNNIRKIIAKRLLESKQTVPHFYLSIECNVDKLLDVREDINKSFSE 231 Query: 712 QRGLVLSVNDFIVKAASLALRLVPEINNGLNADGTIKINHNIDISVAVASESGLITPIVF 891 + +SVNDFI+ A + AL+ VP N + D I+ +N+DISVAVA E+G++TPIV Sbjct: 232 DKVTKISVNDFIILAVAKALQEVPNANASWSEDA-IRYYNNVDISVAVAIENGIVTPIVK 290 Query: 892 GADKLPLAAISSTIHELAEKARNNKLQPQEFMGGTFTISNLGMFGIKEFTAIINSPQAAI 1071 A+K + +S + L +KA++NKL P EF GG FTISNLGM+GIK F AIIN+PQ+ I Sbjct: 291 DANKKNIIELSREMKTLIKKAKDNKLTPIEFQGGGFTISNLGMYGIKNFNAIINTPQSCI 350 Query: 1072 LAVGAGIPHLAHDPANPSKPKSVTKMRFTLCADARIIDETSASEFVSRVKSYLENPSFLL 1251 + VGA + T M TL AD R+ID ++EF++ K ++ENP +L Sbjct: 351 MGVGASTKRAI---VKNDQIIIATIMDVTLSADHRVIDGAVSAEFLASFKRFIENPVLML 407
>sp|P36413|ODP2_DICDI Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (E2) (Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex) (PDC-E2) Length = 592 Score = 234 bits (596), Expect = 5e-61 Identities = 157/431 (36%), Positives = 228/431 (52%), Gaps = 17/431 (3%) Frame = +1 Query: 10 LIKMPSLSPTMSEGGIVEWYKKEGDKISAGDLICDVQTDKAVVGME-ADEEGILAKILKA 186 ++ MP+LSP+M GGI W KKEGD+I AGD I +V+TDKA + + D G LAKIL Sbjct: 165 VVGMPALSPSMETGGIASWTKKEGDQIKAGDAIAEVETDKATMDFQYEDGNGYLAKILVP 224 Query: 187 KDSQGIQVGEAIALLAGEDEDWK-----TVEIPAGGATVGVXXXXXXXXXXXXXXXXXXX 351 + GIQ+ + + ++ ED +VE + ++ Sbjct: 225 GGTSGIQINQPVCIIVKNKEDCDKFADYSVEEQSSSSSSS-SQESTPSSSSSSSQESTPS 283 Query: 352 XRKSNKATSKLG------PAVRRLLDTYGINENQVDHSGPHDVLTKSDVLQHIDRSR--L 507 S + T K G PA R + G + + ++ +GP++ + K+DVL+ + + + Sbjct: 284 QSSSQQTTRKSGERIFATPAARFEASSKGYDLSAINGTGPNNRILKADVLEFVPQKQEVA 343 Query: 508 SPVCPPTTRPAAQPEXXXXXXH-VDVPLTNMRQTIARRMAASKSSIPHRYSTAEASXXXX 684 TT +P D+P +N+R+ A R+ SK +IPH Y T E Sbjct: 344 QQQQQQTTTTTKKPTTPTSSGEFTDIPHSNIRKVTAARLTESKQTIPHYYLTMECRVDKL 403 Query: 685 XXXXXXXXDQRGLVLSVNDFIVKAASLALRLVPEINNGLNADGTIKINHNIDISVAVASE 864 + +SVNDFIVKA+ ALR P +N+ D I+ HNIDI+VAV + Sbjct: 404 LKLRSELNAMNTVKISVNDFIVKASLPALRDNPVVNSTWT-DQFIRRYHNIDINVAVNTP 462 Query: 865 SGLITPIVFGADKLPLAAISSTIHELAEKARNNKLQPQEFMGGTFTISNLGMFGIKEFTA 1044 GL TPIV G D L +IS+++ +LAEKA+N KL P EF GTFTISNLGM GIK+F A Sbjct: 463 QGLFTPIVRGVDMKGLNSISTSVKQLAEKAQNGKLHPSEFESGTFTISNLGMLGIKQFAA 522 Query: 1045 IINSPQAAILAVGAG--IPHLAHDPANPSKPKSVTKMRFTLCADARIIDETSASEFVSRV 1218 +IN PQAAILA+ + L++ P +P ++ T + TL D R+ID +E++ Sbjct: 523 VINPPQAAILALVPQKLVSFLSNKPDSPY--ETATILSVTLSCDHRVIDGAVGAEWLKSF 580 Query: 1219 KSYLENPSFLL 1251 K Y+ENP L+ Sbjct: 581 KDYVENPIKLI 591
Score = 82.0 bits (201), Expect = 3e-15
Identities = 39/79 (49%), Positives = 57/79 (72%)
Frame = +1
Query: 13 IKMPSLSPTMSEGGIVEWYKKEGDKISAGDLICDVQTDKAVVGMEADEEGILAKILKAKD 192
I MP+LSP+M+ G IV+W KKEGD+I AGD+I +V+TDKA + D G LAKIL +
Sbjct: 44 ITMPALSPSMTVGNIVQWKKKEGDQIKAGDVIREVETDKATMDSYEDGNGYLAKILIPEG 103
Query: 193 SQGIQVGEAIALLAGEDED 249
++GI++ + IA++ + ED
Sbjct: 104 TKGIEINKPIAIIVSKKED 122
>sp|Q92HK7|ODP2_RICCN Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex (E2) (Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex) Length = 412 Score = 229 bits (585), Expect = 1e-59 Identities = 148/422 (35%), Positives = 222/422 (52%), Gaps = 9/422 (2%) Frame = +1 Query: 13 IKMPSLSPTMSEGGIVEWYKKEGDKISAGDLICDVQTDKAVVGMEADEEGILAKILKAKD 192 I MP+LSPTM+EG + W KKEGDK++ G++I +++TDKA + +EA +EGILAKI+ ++ Sbjct: 5 ILMPALSPTMTEGNLARWLKKEGDKVNPGEVIAEIETDKATMEVEAVDEGILAKIVIPQN 64 Query: 193 SQGIQVGEAIALLAGEDEDWKTVE-IPAGGATVGVXXXXXXXXXXXXXXXXXXXXR---- 357 SQ + V IA+L+ E E+ ++ A +V + Sbjct: 65 SQNVPVNSLIAVLSEEGEEKTDIDAFIAKNNSVSPSPKTDANLPKPHENIANVEEQVTVI 124 Query: 358 KSNKATSKLGPAVRRLLDTYGINENQVDHSGPHDVLTKSDVLQHIDRSRLSPVCPPTTRP 537 K + + P +RL I V SGPH + K D+L + + + + Sbjct: 125 KHDVSRIFASPLAKRLAKMRNIRFESVKGSGPHGRIVKQDILSYTPSTAHNKIV------ 178 Query: 538 AAQPEXXXXXXHVDVPLTNMRQTIARRMAASKSSIPHRYSTAEASXXXXXXXXXXXX--- 708 + PE VP N+R+ IA+R+ SK ++PH Y + E + Sbjct: 179 SRNPEEYRL-----VPNNNIRKIIAKRLLESKQTVPHFYLSIECNVDKLLDIREDINKFF 233 Query: 709 -DQRGLVLSVNDFIVKAASLALRLVPEINNGLNADGTIKINHNIDISVAVASESGLITPI 885 + + +SVNDFI+ A + AL+ VP N D I+ +N+DISVAVA E+GL+TPI Sbjct: 234 SEDKSTRISVNDFIILAVAKALQEVPNANASWGEDA-IRYYNNVDISVAVAIENGLVTPI 292 Query: 886 VFGADKLPLAAISSTIHELAEKARNNKLQPQEFMGGTFTISNLGMFGIKEFTAIINSPQA 1065 V A++ + +S + L +KA++NKL P+EF GG FTISNLGM+GIK F AIIN PQ+ Sbjct: 293 VKNANQKNILELSREMKALIKKAKDNKLTPEEFQGGGFTISNLGMYGIKNFNAIINPPQS 352 Query: 1066 AILAVGAGIPHLAHDPANPSKPKSVTKMRFTLCADARIIDETSASEFVSRVKSYLENPSF 1245 I+ VGA + T M TL AD R++D +EF+ K ++E+P Sbjct: 353 CIMGVGASAKRAI---VKNDQITIATIMDVTLSADHRVVDGAVGAEFLVAFKKFIESPVL 409 Query: 1246 LL 1251 +L Sbjct: 410 ML 411
>sp|Q9R9N3|ODP2_RHIME Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex (E2) (Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex) Length = 447 Score = 225 bits (574), Expect = 2e-58 Identities = 151/446 (33%), Positives = 220/446 (49%), Gaps = 33/446 (7%) Frame = +1 Query: 13 IKMPSLSPTMSEGGIVEWYKKEGDKISAGDLICDVQTDKAVVGMEADEEGILAKILKAKD 192 I MP+LSPTM EG + +W KEGDK+ +GD+I +++TDKA + +EA +EG +AKI+ Sbjct: 5 ITMPALSPTMEEGNLAKWLVKEGDKVKSGDVIAEIETDKATMEVEAVDEGTVAKIVVPAG 64 Query: 193 SQGIQVGEAIALLAGEDEDWKTVEIPAGGATVGVXXXXXXXXXXXXXXXXXXXXRK-SNK 369 ++G++V IA+LA E ED T GA V + + Sbjct: 65 TEGVKVNALIAVLAAEGEDVATAAKGGNGAAGAVPAPKPKETAETAPAAAPAPAAAPAPQ 124 Query: 370 ATSKLGPA--------------VRRLLDTYGINENQVDHSGPHDVLTKSDVLQHIDRSRL 507 A + PA RRL GI+ + + SGPH + K DV + Sbjct: 125 AAAPASPAPADGEGKRIFSSPLARRLAKEAGIDLSAIAGSGPHGRVVKKDVETAVSGGAA 184 Query: 508 SPVCPPTTRPA-----------AQPEXXXXXXHVDVPLTNMRQTIARRMAASKSSIPHRY 654 P P PA A + + VP MR+TIA+R+ SK +IPH Y Sbjct: 185 KPAGAPAAAPAPATLAKGMSEDAVLKLFEPGSYELVPHDGMRKTIAKRLVESKQTIPHFY 244 Query: 655 STAE----ASXXXXXXXXXXXXDQRG---LVLSVNDFIVKAASLALRLVPEINNGLNADG 813 + + A ++ G LSVND ++KA +LALR VP+ N Sbjct: 245 VSVDCELDALMALRAQLNAAAPEKDGKPVYKLSVNDMVIKALALALRDVPDANVSWTDQN 304 Query: 814 TIKINHNIDISVAVASESGLITPIVFGADKLPLAAISSTIHELAEKARNNKLQPQEFMGG 993 +K H D+ VAV+ GLITPIV A+ L+AIS+ + +L ++A+ KL+P+E+ GG Sbjct: 305 MVKHKH-ADVGVAVSIPGGLITPIVRQAELKSLSAISNEMKDLGKRAKERKLKPEEYQGG 363 Query: 994 TFTISNLGMFGIKEFTAIINSPQAAILAVGAGIPHLAHDPANPSKPKSVTKMRFTLCADA 1173 T +SN+GM G+K+F A++N P A ILAVGAG + + M TL D Sbjct: 364 TTAVSNMGMMGVKDFAAVVNPPHATILAVGAGEDRVV---VRNKEMVIANVMTVTLSTDH 420 Query: 1174 RIIDETSASEFVSRVKSYLENPSFLL 1251 R +D +E ++ K Y+ENP +L Sbjct: 421 RCVDGALGAELLAAFKRYIENPMGML 446
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.317 0.133 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 151,591,759
Number of Sequences: 369166
Number of extensions: 3163100
Number of successful extensions: 10120
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9360
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10008
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 15276346370
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail