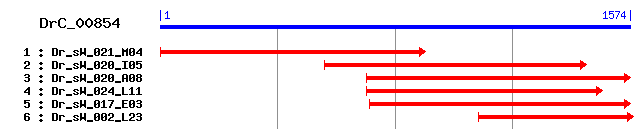
DrC_00854
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00854 (1574 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9UG63|ABCF2_HUMAN ATP-binding cassette sub-family F mem... 680 0.0 sp|Q99LE6|ABCF2_MOUSE ATP-binding cassette sub-family F mem... 679 0.0 sp|P40024|YEM6_YEAST Probable ATP-dependent transporter YER... 463 e-130 sp|P43535|GCN20_YEAST Protein GCN20 360 5e-99 sp|Q6MG08|ABCF1_RAT ATP-binding cassette sub-family F membe... 358 2e-98 sp|Q6P542|ABCF1_MOUSE ATP-binding cassette sub-family F mem... 356 9e-98 sp|Q767L0|ABCF1_PIG ATP-binding cassette sub-family F member 1 355 2e-97 sp|Q8NE71|ABCF1_HUMAN ATP-binding cassette sub-family F mem... 355 3e-97 sp|Q7YR37|ABCF1_PANTR ATP-binding cassette sub-family F mem... 355 3e-97 sp|P44808|Y658_HAEIN Probable ABC transporter ATP-binding p... 288 2e-77
>sp|Q9UG63|ABCF2_HUMAN ATP-binding cassette sub-family F member 2 (Iron-inhibited ABC transporter 2) Length = 623 Score = 680 bits (1755), Expect = 0.0 Identities = 325/499 (65%), Positives = 398/499 (79%) Frame = +2 Query: 5 NGCGKSTLFAVLSNNELPIPSHIDQFLLQREMPPSDKTALQAVMEADVEKIRLEKEAEYL 184 NG GKS L + + E+PIP HID + L REMPPSDKT L VME D E+ LEKEAE L Sbjct: 120 NGIGKSMLLSAIGKREVPIPEHIDIYHLTREMPPSDKTPLHCVMEVDTERAMLEKEAERL 179 Query: 185 ALQDDTESQDRLIEVYERLXXXXXXXXXXXXXXXXFGLGFTKDMQNKEVKHFSGGWRMRI 364 A +D ++L+E+YERL GLGFT MQ K++K FSGGWRMR+ Sbjct: 180 AHED--AECEKLMELYERLEELDADKAEMRASRILHGLGFTPAMQRKKLKDFSGGWRMRV 237 Query: 365 SLARALFVKPALLMLDEPTNHLDLNACVWLEKELSQYNRILLIISHSEDFLNGVCTNIVH 544 +LARALF++P +L+LDEPTNHLDL+ACVWLE+EL + RIL+++SHS+DFLNGVCTNI+H Sbjct: 238 ALARALFIRPFMLLLDEPTNHLDLDACVWLEEELKTFKRILVLVSHSQDFLNGVCTNIIH 297 Query: 545 MQNKKLVYYGGNYDQYIMTKMELEENQMKKYKWEQDQMAHMKDYIARFGHGSRKLARQAQ 724 M NKKL YY GNYDQY+ T++ELEENQMK++ WEQDQ+AHMK+YIARFGHGS KLARQAQ Sbjct: 298 MHNKKLKYYTGNYDQYVKTRLELEENQMKRFHWEQDQIAHMKNYIARFGHGSAKLARQAQ 357 Query: 725 SKEKTLKKMVDGGLTQKVIREKNLVFSFPSCGEIPPPVIQVQNVSFKYGANKPLIYDNID 904 SKEKTL+KM+ GLT++V+ +K L F FP CG+IPPPVI VQNVSFKY + P IY+N++ Sbjct: 358 SKEKTLQKMMASGLTERVVSDKTLSFYFPPCGKIPPPVIMVQNVSFKYTKDGPCIYNNLE 417 Query: 905 FGVDLDTRVALVGPNGAGKSTLLKLIASELEPTDGLIRRHSKCRIGRYHQHLHEILDLNS 1084 FG+DLDTRVALVGPNGAGKSTLLKL+ EL PTDG+IR+HS +IGRYHQHL E LDL+ Sbjct: 418 FGIDLDTRVALVGPNGAGKSTLLKLLTGELLPTDGMIRKHSHVKIGRYHQHLQEQLDLDL 477 Query: 1085 TALEWMMKSFPHIKEKDEMRKKLGRYGLTGGQQICPIKSLSDGQRCRIIFAWLAEGCPHM 1264 + LE+MMK +P IKEK+EMRK +GRYGLTG QQ+ PI++LSDGQ+CR+ AWLA PHM Sbjct: 478 SPLEYMMKCYPEIKEKEEMRKIIGRYGLTGKQQVSPIRNLSDGQKCRVCLAWLAWQNPHM 537 Query: 1265 LLLDEPTNHLDLETIDSLADAINEFEGGLLLVSHDFRLIGRVAEEIWVCDHGAVTKWPGD 1444 L LDEPTNHLD+ETID+LADAINEFEGG++LVSHDFRLI +VA+EIWVC+ +TKWPGD Sbjct: 538 LFLDEPTNHLDIETIDALADAINEFEGGMMLVSHDFRLIQQVAQEIWVCEKQTITKWPGD 597 Query: 1445 IYSYKTSLIKEIEREHTRM 1501 I +YK L ++ E ++ Sbjct: 598 ILAYKEHLKSKLVDEEPQL 616
Score = 60.8 bits (146), Expect = 1e-08
Identities = 63/221 (28%), Positives = 89/221 (40%), Gaps = 25/221 (11%)
Frame = +2
Query: 926 RVALVGPNGAGKSTLLKLIAS----------------ELEPTDGLIRRHSKCRIGRYHQH 1057
R L+G NG GKS LL I E+ P+D H +
Sbjct: 113 RYGLIGLNGIGKSMLLSAIGKREVPIPEHIDIYHLTREMPPSDKT-PLHCVMEVDTERAM 171
Query: 1058 LH---EILDLNSTALEWMMKSFPHIKEKD----EMRKK--LGRYGLTGGQQICPIKSLSD 1210
L E L E +M+ + ++E D EMR L G T Q +K S
Sbjct: 172 LEKEAERLAHEDAECEKLMELYERLEELDADKAEMRASRILHGLGFTPAMQRKKLKDFSG 231
Query: 1211 GQRCRIIFAWLAEGCPHMLLLDEPTNHLDLETIDSLADAINEFEGGLLLVSHDFRLIGRV 1390
G R R+ A P MLLLDEPTNHLDL+ L + + F+ L+LVSH + V
Sbjct: 232 GWRMRVALARALFIRPFMLLLDEPTNHLDLDACVWLEEELKTFKRILVLVSHSQDFLNGV 291
Query: 1391 AEEIWVCDHGAVTKWPGDIYSYKTSLIKEIEREHTRMMTER 1513
I + + + G+ Y + ++ E + R E+
Sbjct: 292 CTNIIHMHNKKLKYYTGNYDQYVKTRLELEENQMKRFHWEQ 332
>sp|Q99LE6|ABCF2_MOUSE ATP-binding cassette sub-family F member 2 Length = 628 Score = 679 bits (1753), Expect = 0.0 Identities = 324/499 (64%), Positives = 399/499 (79%) Frame = +2 Query: 5 NGCGKSTLFAVLSNNELPIPSHIDQFLLQREMPPSDKTALQAVMEADVEKIRLEKEAEYL 184 NG GKS L + + E+PIP HID + L REMPPS+KT LQ VME D E+ LE+EAE L Sbjct: 125 NGIGKSMLLSAIGKREVPIPEHIDIYHLTREMPPSEKTPLQCVMEVDTERAMLEREAERL 184 Query: 185 ALQDDTESQDRLIEVYERLXXXXXXXXXXXXXXXXFGLGFTKDMQNKEVKHFSGGWRMRI 364 A +D ++L+E+YERL GLGFT MQ K++K FSGGWRMR+ Sbjct: 185 AHED--AECEKLMELYERLEELDADKAEMRASRILHGLGFTPAMQRKKLKDFSGGWRMRV 242 Query: 365 SLARALFVKPALLMLDEPTNHLDLNACVWLEKELSQYNRILLIISHSEDFLNGVCTNIVH 544 +LARALF++P +L+LDEPTNHLDL+ACVWLE+EL + RIL+++SHS+DFLNGVCTNI+H Sbjct: 243 ALARALFIRPFMLLLDEPTNHLDLDACVWLEEELKTFKRILVLVSHSQDFLNGVCTNIIH 302 Query: 545 MQNKKLVYYGGNYDQYIMTKMELEENQMKKYKWEQDQMAHMKDYIARFGHGSRKLARQAQ 724 M NKKL YY GNYDQY+ T++ELEENQMK++ WEQDQ+AHMK+YIARFGHGS KLARQAQ Sbjct: 303 MHNKKLKYYTGNYDQYVKTRLELEENQMKRFHWEQDQIAHMKNYIARFGHGSAKLARQAQ 362 Query: 725 SKEKTLKKMVDGGLTQKVIREKNLVFSFPSCGEIPPPVIQVQNVSFKYGANKPLIYDNID 904 SKEKTL+KM+ GLT++V+ +K L F FP CG+IPPPVI VQNVSFKY + P IY+N++ Sbjct: 363 SKEKTLQKMMASGLTERVVSDKTLSFYFPPCGKIPPPVIMVQNVSFKYTKDGPCIYNNLE 422 Query: 905 FGVDLDTRVALVGPNGAGKSTLLKLIASELEPTDGLIRRHSKCRIGRYHQHLHEILDLNS 1084 FG+DLDTRVALVGPNGAGKSTLLKL+ EL PTDG+IR+HS +IGRYHQHL E LDL+ Sbjct: 423 FGIDLDTRVALVGPNGAGKSTLLKLLTGELLPTDGMIRKHSHVKIGRYHQHLQEQLDLDL 482 Query: 1085 TALEWMMKSFPHIKEKDEMRKKLGRYGLTGGQQICPIKSLSDGQRCRIIFAWLAEGCPHM 1264 + LE+MMK +P IKEK+EMRK +GRYGLTG QQ+ PI++LSDGQ+CR+ AWLA PHM Sbjct: 483 SPLEYMMKCYPEIKEKEEMRKIIGRYGLTGKQQVSPIRNLSDGQKCRVCLAWLAWQNPHM 542 Query: 1265 LLLDEPTNHLDLETIDSLADAINEFEGGLLLVSHDFRLIGRVAEEIWVCDHGAVTKWPGD 1444 L LDEPTNHLD+ETID+LADAINEFEGG++LVSHDFRLI +VA+EIWVC+ +TKWPGD Sbjct: 543 LFLDEPTNHLDIETIDALADAINEFEGGMMLVSHDFRLIQQVAQEIWVCEKQTITKWPGD 602 Query: 1445 IYSYKTSLIKEIEREHTRM 1501 I +YK L ++ E ++ Sbjct: 603 ILAYKEHLKSKLVDEEPQL 621
Score = 58.2 bits (139), Expect = 7e-08
Identities = 62/220 (28%), Positives = 87/220 (39%), Gaps = 24/220 (10%)
Frame = +2
Query: 926 RVALVGPNGAGKSTLLKLIASELEPTDGLIRRHSKCR---------------IGRYHQHL 1060
R L+G NG GKS LL I P I + R + L
Sbjct: 118 RYGLIGLNGIGKSMLLSAIGKREVPIPEHIDIYHLTREMPPSEKTPLQCVMEVDTERAML 177
Query: 1061 H---EILDLNSTALEWMMKSFPHIKEKD----EMRKK--LGRYGLTGGQQICPIKSLSDG 1213
E L E +M+ + ++E D EMR L G T Q +K S G
Sbjct: 178 EREAERLAHEDAECEKLMELYERLEELDADKAEMRASRILHGLGFTPAMQRKKLKDFSGG 237
Query: 1214 QRCRIIFAWLAEGCPHMLLLDEPTNHLDLETIDSLADAINEFEGGLLLVSHDFRLIGRVA 1393
R R+ A P MLLLDEPTNHLDL+ L + + F+ L+LVSH + V
Sbjct: 238 WRMRVALARALFIRPFMLLLDEPTNHLDLDACVWLEEELKTFKRILVLVSHSQDFLNGVC 297
Query: 1394 EEIWVCDHGAVTKWPGDIYSYKTSLIKEIEREHTRMMTER 1513
I + + + G+ Y + ++ E + R E+
Sbjct: 298 TNIIHMHNKKLKYYTGNYDQYVKTRLELEENQMKRFHWEQ 337
>sp|P40024|YEM6_YEAST Probable ATP-dependent transporter YER036C Length = 610 Score = 463 bits (1191), Expect = e-130 Identities = 239/497 (48%), Positives = 334/497 (67%), Gaps = 4/497 (0%) Frame = +2 Query: 2 QNGCGKSTLFAVLSNNELPIPSHIDQFLLQREMPPSDKTALQAVM-EADVEKIRLEKEAE 178 +NGCGKST L+ E PIP HID +LL PS+ +AL V+ EA E R+E E Sbjct: 115 ENGCGKSTFLKALATREYPIPEHIDIYLLDEPAEPSELSALDYVVTEAQHELKRIEDLVE 174 Query: 179 YLALQDDTESQDRLIE-VYERLXXXXXXXXXXXXXXXXFGLGFTKDMQNKEVKHFSGGWR 355 L+D ES+ L+E +YER+ GLGF K K+ K SGGW+ Sbjct: 175 KTILEDGPESE--LLEPLYERMDSLDPDTFESRAAIILIGLGFNKKTILKKTKDMSGGWK 232 Query: 356 MRISLARALFVKPALLMLDEPTNHLDLNACVWLEKELSQYNRILLIISHSEDFLNGVCTN 535 MR++LA+ALFVKP LL+LD+PT HLDL ACVWLE+ L +++R L+++SHS+DFLNGVCTN Sbjct: 233 MRVALAKALFVKPTLLLLDDPTAHLDLEACVWLEEYLKRFDRTLVLVSHSQDFLNGVCTN 292 Query: 536 IVHMQNKKLVYYGGNYDQYIMTKMELEENQMKKYKWEQDQMAHMKDYIARFGHGSRKLAR 715 ++ M+ +KL YGGNYD Y T+ ELE NQMK+Y +Q+++ H+K +IA G + L + Sbjct: 293 MIDMRAQKLTAYGGNYDSYHKTRSELETNQMKQYNKQQEEIQHIKKFIASAGTYA-NLVK 351 Query: 716 QAQSKEKTLKKMVDGGLTQKVIREKNLVFSFPSCGEIPPPVIQVQNVSFKYGAN-KPLIY 892 QA+S++K L KM GL Q V+ +K F FP +PPPV+ ++SF Y +N +Y Sbjct: 352 QAKSRQKILDKMEADGLVQPVVPDKVFSFRFPQVERLPPPVLAFDDISFHYESNPSENLY 411 Query: 893 DNIDFGVDLDTRVALVGPNGAGKSTLLKLIASELEPTDGLIRRHSKCRIGRYHQHLHEIL 1072 ++++FGVD+D+R+ALVGPNG GKSTLLK++ EL P G + RH+ ++G Y QH + L Sbjct: 412 EHLNFGVDMDSRIALVGPNGVGKSTLLKIMTGELTPQSGRVSRHTHVKLGVYSQHSQDQL 471 Query: 1073 DLNSTALEWMMKSFPHIKEKDEM-RKKLGRYGLTGGQQICPIKSLSDGQRCRIIFAWLAE 1249 DL +ALE++ + +I + + R +LGRYGLTG Q + +LS+GQR R++FA LA Sbjct: 472 DLTKSALEFVRDKYSNISQDFQFWRGQLGRYGLTGEGQTVQMATLSEGQRSRVVFALLAL 531 Query: 1250 GCPHMLLLDEPTNHLDLETIDSLADAINEFEGGLLLVSHDFRLIGRVAEEIWVCDHGAVT 1429 P++LLLDEPTN LD+ TIDSLADAINEF GG+++VSHDFRL+ ++A++I+V ++ T Sbjct: 532 EQPNVLLLDEPTNGLDIPTIDSLADAINEFNGGVVVVSHDFRLLDKIAQDIFVVENKTAT 591 Query: 1430 KWPGDIYSYKTSLIKEI 1480 +W G I YK L K + Sbjct: 592 RWDGSILQYKNKLAKNV 608
>sp|P43535|GCN20_YEAST Protein GCN20 Length = 752 Score = 360 bits (925), Expect = 5e-99 Identities = 195/518 (37%), Positives = 312/518 (60%), Gaps = 24/518 (4%) Frame = +2 Query: 2 QNGCGKSTLFAVLSNNELPIPSHIDQFLLQREMPPSDKTALQAVMEADV----------- 148 QNG GKSTL LS EL +P H+ +++E+ D ALQ+V++ADV Sbjct: 233 QNGIGKSTLLRALSRRELNVPKHVSILHVEQELRGDDTKALQSVLDADVWRKQLLSEEAK 292 Query: 149 --------EKIRLEKEAEYLALQD-DTESQD---RLIEVYERLXXXXXXXXXXXXXXXXF 292 + +R E E + L ++ D E +D LI++ ++L + Sbjct: 293 INERLKEMDVLRQEFEEDSLEVKKLDNEREDLDNHLIQISDKLVDMESDKAEARAASILY 352 Query: 293 GLGFTKDMQNKEVKHFSGGWRMRISLARALFVKPALLMLDEPTNHLDLNACVWLEKELSQ 472 GLGF+ + Q + FSGGWRMR+SLARALF +P LL+LDEP+N LD+ + +L + L Sbjct: 353 GLGFSTEAQQQPTNSFSGGWRMRLSLARALFCQPDLLLLDEPSNMLDVPSIAYLAEYLKT 412 Query: 473 YNRILLIISHSEDFLNGVCTNIVHMQNKKLVYYGG-NYDQYIMTKMELEENQMKKYKWEQ 649 Y +L +SH FLN V T+I++ N++L YY G ++D + TK E +N ++Y + Sbjct: 413 YPNTVLTVSHDRAFLNEVATDIIYQHNERLDYYRGQDFDTFYTTKEERRKNAQREYDNQM 472 Query: 650 DQMAHMKDYIARFGHGSRKLARQAQSKEKTLKKMVDGGLTQKVIREKNLVFSFPSCGEIP 829 H++++I ++ + + K +++AQS+ K L+K+ + + ++K + F FP C ++ Sbjct: 473 VYRKHLQEFIDKYRYNAAK-SQEAQSRIKKLEKLP---VLEPPEQDKTIDFKFPECDKLS 528 Query: 830 PPVIQVQNVSFKYGANKPLIYDNIDFGVDLDTRVALVGPNGAGKSTLLKLIASELEPTDG 1009 PP+IQ+Q+VSF Y N L+ D ++ V +D+R+ALVG NG GK+TLLK++ +L P G Sbjct: 529 PPIIQLQDVSFGYDENNLLLKD-VNLDVQMDSRIALVGANGCGKTTLLKIMMEQLRPLKG 587 Query: 1010 LIRRHSKCRIGRYHQHLHEILDLNSTALEWMMKSFPHIKEKDEMRKKLGRYGLTGGQQIC 1189 + R+ + RIG + QH + +DL ++A++WM KSFP K +E R+ LG +G+TG + Sbjct: 588 FVSRNPRLRIGYFTQHHVDSMDLTTSAVDWMSKSFPG-KTDEEYRRHLGSFGITGTLGLQ 646 Query: 1190 PIKSLSDGQRCRIIFAWLAEGCPHMLLLDEPTNHLDLETIDSLADAINEFEGGLLLVSHD 1369 ++ LS GQ+ R+ FA L PH+L+LDEP+NHLD +D+L +A+ F GG+L+VSHD Sbjct: 647 KMQLLSGGQKSRVAFAALCLNNPHILVLDEPSNHLDTTGLDALVEALKNFNGGVLMVSHD 706 Query: 1370 FRLIGRVAEEIWVCDHGAVTKWPGDIYSYKTSLIKEIE 1483 +I V +EIWV + G V ++ G IY Y+ +++ + Sbjct: 707 ISVIDSVCKEIWVSEQGTVKRFEGTIYDYRDYILQSAD 744
>sp|Q6MG08|ABCF1_RAT ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) Length = 839 Score = 358 bits (919), Expect = 2e-98 Identities = 200/509 (39%), Positives = 305/509 (59%), Gaps = 17/509 (3%) Frame = +2 Query: 5 NGCGKSTLFAVLSNNELPIPSHIDQFLLQREMPPSDKTALQAVMEADVEKIRL---EKEA 175 NG GK+TL ++N L IP +ID L ++E+ + A+QAV+ AD +++RL EK Sbjct: 332 NGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQAVLRADTKRLRLLEEEKRL 391 Query: 176 EYLALQDDTESQDRLIEVYERLXXXXXXXXXXXXXXXXFGLGFTKDMQNKEVKHFSGGWR 355 + Q D + ++L +VYE L GLGF +MQN+ + FSGGWR Sbjct: 392 QGQLEQGDDTAAEKLEKVYEELRATGAAAAEAKARRILAGLGFDPEMQNRPTQKFSGGWR 451 Query: 356 MRISLARALFVKPALLMLDEPTNHLDLNACVWLEKELSQYNRILLIISHSEDFLNGVCTN 535 MR+SLARALF++P LLMLDEPTNHLDLNA +WL L + + LLI+SH + FL+ VCT+ Sbjct: 452 MRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWRKTLLIVSHDQGFLDDVCTD 511 Query: 536 IVHMQNKKLVYYGGNYDQYIMTKMELEENQMKKYKWEQDQMAHMKDYIARFGHGSRKLAR 715 I+H+ ++L YY GNY + + ++ +K+Y+ ++ ++ +K G S K A Sbjct: 512 IIHLDTQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKLKELK-----AGGKSTKQA- 565 Query: 716 QAQSKEKTLKKM------------VDGGLTQKVIREKNLVFSFPSCGEIPPPVIQVQNVS 859 + Q+KE +K D K RE + F+FP + PPV+ + V+ Sbjct: 566 EKQTKEVLTRKQQKCRRKNQDEESQDPPELLKRpreYTVRFTFPDPPPLSPPVLGLHGVT 625 Query: 860 FKYGANKPLIYDNIDFGVDLDTRVALVGPNGAGKSTLLKLIASELEPTDGLIRRHSKCRI 1039 F Y KPL + N+DFG+D+D+R+ +VGPNG GKSTLL L+ +L PT+G +R++ + +I Sbjct: 626 FGYEGQKPL-FKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLTPTNGEMRKNHRLKI 684 Query: 1040 GRYHQHLHEILDLNSTALEWMMKSF--PHIKEKDEMRKKLGRYGLTGGQQICPIKSLSDG 1213 G ++Q E L + T E++ + F P+ + RK LGR+GL I LS G Sbjct: 685 GFFNQQYAEQLHMEETPTEYLQRGFNLPY----QDARKCLGRFGLESHAHTIQICKLSGG 740 Query: 1214 QRCRIIFAWLAEGCPHMLLLDEPTNHLDLETIDSLADAINEFEGGLLLVSHDFRLIGRVA 1393 Q+ R++FA LA P +L+LDEPTN+LD+E+ID+L +AINE++G +++VSHD RLI Sbjct: 741 QKARVVFAELACREPDVLILDEPTNNLDIESIDALGEAINEYKGAVIVVSHDARLITETN 800 Query: 1394 EEIWVCDHGAVTKWPGDIYSYKTSLIKEI 1480 ++WV + +V++ GD YK +++ + Sbjct: 801 CQLWVVEEQSVSQIDGDFDDYKREVLEAL 829
Score = 74.7 bits (182), Expect = 7e-13
Identities = 78/309 (25%), Positives = 132/309 (42%), Gaps = 45/309 (14%)
Frame = +2
Query: 710 ARQAQSKEKTLKKMVDGGLTQKVIREKNLV---FSFPSCGEIPPPVIQVQNVS------F 862
A ++ ++K LKK +D + ++ N FS S E+ ++N S F
Sbjct: 245 AHLSKKEKKKLKKQMDYERQVESLKAANAAENDFSV-SQAEVSSRQAMLENASDIKLEKF 303
Query: 863 KYGANKPLIYDNIDFGVDLDTRVALVGPNGAGKSTLLK---------------------L 979
A+ ++ N D + R LVGPNG GK+TLLK +
Sbjct: 304 SISAHGKELFVNADLYIVAGRRYGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEV 363
Query: 980 IASELEPTDGLIRRHSK-CRIGRYHQHLHEILDL-NSTALEWMMKSFPHIK------EKD 1135
+A E ++R +K R+ + L L+ + TA E + K + ++ +
Sbjct: 364 VADETPAVQAVLRADTKRLRLLEEEKRLQGQLEQGDDTAAEKLEKVYEELRATGAAAAEA 423
Query: 1136 EMRKKLGRYGLTGGQQICPIKSLSDGQRCRIIFAWLAEGCPHMLLLDEPTNHLDLETIDS 1315
+ R+ L G Q P + S G R R+ A P +L+LDEPTNHLDL +
Sbjct: 424 KARRILAGLGFDPEMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVIW 483
Query: 1316 LADAINEFEGGLLLVSHDFRLIGRVAEEIWVCDHGAVTKWPGDIYSYK-------TSLIK 1474
L + + + LL+VSHD + V +I D + + G+ ++K L+K
Sbjct: 484 LNNYLQGWRKTLLIVSHDQGFLDDVCTDIIHLDTQRLHYYRGNYMTFKKMYQQKQKELLK 543
Query: 1475 EIEREHTRM 1501
+ E++ ++
Sbjct: 544 QYEKQEKKL 552
>sp|Q6P542|ABCF1_MOUSE ATP-binding cassette sub-family F member 1 Length = 837 Score = 356 bits (914), Expect = 9e-98 Identities = 197/508 (38%), Positives = 309/508 (60%), Gaps = 16/508 (3%) Frame = +2 Query: 5 NGCGKSTLFAVLSNNELPIPSHIDQFLLQREMPPSDKTALQAVMEADVEKIRLEKEAEYL 184 NG GK+TL ++N L IP +ID L ++E+ + A+QAV+ AD +++RL +E L Sbjct: 330 NGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQAVLRADTKRLRLLEEERRL 389 Query: 185 ALQ----DDTESQDRLIEVYERLXXXXXXXXXXXXXXXXFGLGFTKDMQNKEVKHFSGGW 352 Q DDT ++ +L +VYE L GLGF +MQN+ + FSGGW Sbjct: 390 QGQLEQGDDTAAE-KLEKVYEELRATGAAAAEAKARRILAGLGFDPEMQNRPTQKFSGGW 448 Query: 353 RMRISLARALFVKPALLMLDEPTNHLDLNACVWLEKELSQYNRILLIISHSEDFLNGVCT 532 RMR+SLARALF++P LLMLDEPTNHLDLNA +WL L + + LLI+SH + FL+ VCT Sbjct: 449 RMRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWRKTLLIVSHDQGFLDDVCT 508 Query: 533 NIVHMQNKKLVYYGGNYDQYIMTKMELEENQMKKYKWEQDQMAHMKDYIARFGHGSRKLA 712 +I+H+ ++L YY GNY + + ++ +K+Y+ ++ ++ +K G +++ Sbjct: 509 DIIHLDTQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKLKELK----AGGKSTKQAE 564 Query: 713 RQ-----AQSKEKTLKKMVDGGLTQ-----KVIREKNLVFSFPSCGEIPPPVIQVQNVSF 862 +Q + ++K +K D + K +E + F+FP + PPV+ + V+F Sbjct: 565 KQTKEVLTRKQQKCRRKNQDEESQEPPELLKRPKEYTVRFTFPDPPPLSPPVLGLHGVTF 624 Query: 863 KYGANKPLIYDNIDFGVDLDTRVALVGPNGAGKSTLLKLIASELEPTDGLIRRHSKCRIG 1042 Y KPL + N+DFG+D+D+R+ +VGPNG GKSTLL L+ +L PT+G +R++ + +IG Sbjct: 625 GYEGQKPL-FKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLTPTNGEMRKNHRLKIG 683 Query: 1043 RYHQHLHEILDLNSTALEWMMKSF--PHIKEKDEMRKKLGRYGLTGGQQICPIKSLSDGQ 1216 ++Q E L + T E++ +SF P+ + RK LGR+GL I LS GQ Sbjct: 684 FFNQQYAEQLHMEETPTEYLQRSFNLPY----QDARKCLGRFGLESHAHTIQICKLSGGQ 739 Query: 1217 RCRIIFAWLAEGCPHMLLLDEPTNHLDLETIDSLADAINEFEGGLLLVSHDFRLIGRVAE 1396 + R++FA LA P +L+LDEPTN+LD+E+ID+L +AIN+++G +++VSHD RLI Sbjct: 740 KARVVFAELACREPDVLILDEPTNNLDIESIDALGEAINDYKGAVIVVSHDARLITETNC 799 Query: 1397 EIWVCDHGAVTKWPGDIYSYKTSLIKEI 1480 ++WV + V++ GD YK +++ + Sbjct: 800 QLWVVEEQGVSQIDGDFDDYKREVLEAL 827
Score = 74.7 bits (182), Expect = 7e-13
Identities = 78/309 (25%), Positives = 132/309 (42%), Gaps = 45/309 (14%)
Frame = +2
Query: 710 ARQAQSKEKTLKKMVDGGLTQKVIREKNLV---FSFPSCGEIPPPVIQVQNVS------F 862
A ++ ++K LKK +D + ++ N FS S E+ ++N S F
Sbjct: 243 ANLSKKEKKKLKKQMDYERQVESLKAANAAENDFSV-SQAEVSSRQAMLENASDIKLEKF 301
Query: 863 KYGANKPLIYDNIDFGVDLDTRVALVGPNGAGKSTLLK---------------------L 979
A+ ++ N D + R LVGPNG GK+TLLK +
Sbjct: 302 SISAHGKELFVNADLYIVAGRRYGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEV 361
Query: 980 IASELEPTDGLIRRHSK-CRIGRYHQHLHEILDL-NSTALEWMMKSFPHIK------EKD 1135
+A E ++R +K R+ + L L+ + TA E + K + ++ +
Sbjct: 362 VADETPAVQAVLRADTKRLRLLEEERRLQGQLEQGDDTAAEKLEKVYEELRATGAAAAEA 421
Query: 1136 EMRKKLGRYGLTGGQQICPIKSLSDGQRCRIIFAWLAEGCPHMLLLDEPTNHLDLETIDS 1315
+ R+ L G Q P + S G R R+ A P +L+LDEPTNHLDL +
Sbjct: 422 KARRILAGLGFDPEMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVIW 481
Query: 1316 LADAINEFEGGLLLVSHDFRLIGRVAEEIWVCDHGAVTKWPGDIYSYK-------TSLIK 1474
L + + + LL+VSHD + V +I D + + G+ ++K L+K
Sbjct: 482 LNNYLQGWRKTLLIVSHDQGFLDDVCTDIIHLDTQRLHYYRGNYMTFKKMYQQKQKELLK 541
Query: 1475 EIEREHTRM 1501
+ E++ ++
Sbjct: 542 QYEKQEKKL 550
>sp|Q767L0|ABCF1_PIG ATP-binding cassette sub-family F member 1 Length = 807 Score = 355 bits (912), Expect = 2e-97 Identities = 198/508 (38%), Positives = 308/508 (60%), Gaps = 16/508 (3%) Frame = +2 Query: 5 NGCGKSTLFAVLSNNELPIPSHIDQFLLQREMPPSDKTALQAVMEADVEKIRLEKEAEYL 184 NG GK+TL ++N L IP +ID L ++E+ + A+QAV+ AD ++++L +E L Sbjct: 300 NGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQAVLRADTKRLKLLEEERRL 359 Query: 185 ALQ----DDTESQDRLIEVYERLXXXXXXXXXXXXXXXXFGLGFTKDMQNKEVKHFSGGW 352 Q DDT + DRL +VYE L GLGF +MQN+ + FSGGW Sbjct: 360 QGQLEQGDDTAA-DRLEKVYEELRATGAAAAEAKARRILAGLGFDPEMQNRPTQKFSGGW 418 Query: 353 RMRISLARALFVKPALLMLDEPTNHLDLNACVWLEKELSQYNRILLIISHSEDFLNGVCT 532 RMR+SLARALF++P LLMLDEPTNHLDLNA +WL L + + LLI+SH + FL+ VCT Sbjct: 419 RMRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWRKTLLIVSHDQGFLDDVCT 478 Query: 533 NIVHMQNKKLVYYGGNYDQYIMTKMELEENQMKKYKWEQDQMAHMKDYIARFGHGSRKLA 712 +I+H+ ++L YY GNY + + ++ +K+Y+ ++ ++ +K G +++ Sbjct: 479 DIIHLDAQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKLKELK----AGGKSTKQAE 534 Query: 713 RQ-----AQSKEKTLKKMVDGGLTQ-----KVIREKNLVFSFPSCGEIPPPVIQVQNVSF 862 +Q + ++K +K D + K +E + F+FP + PPV+ + V+F Sbjct: 535 KQTKEALTRKQQKCRRKNQDEESQEAPELLKRPKEYTVRFTFPDPPPLSPPVLGLHGVTF 594 Query: 863 KYGANKPLIYDNIDFGVDLDTRVALVGPNGAGKSTLLKLIASELEPTDGLIRRHSKCRIG 1042 Y KPL + N+DFG+D+D+R+ +VGPNG GKSTLL L+ +L PT G +R++ + +IG Sbjct: 595 GYEGQKPL-FKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLTPTRGEMRKNHRLKIG 653 Query: 1043 RYHQHLHEILDLNSTALEWMMKSF--PHIKEKDEMRKKLGRYGLTGGQQICPIKSLSDGQ 1216 ++Q E L + T E++ + F P+ + RK LGR+GL I LS GQ Sbjct: 654 FFNQQYAEQLRMEETPTEYLQRGFNLPY----QDARKCLGRFGLESHAHTIQICKLSGGQ 709 Query: 1217 RCRIIFAWLAEGCPHMLLLDEPTNHLDLETIDSLADAINEFEGGLLLVSHDFRLIGRVAE 1396 + R++FA LA P +L+LDEPTN+LD+E+ID+L +AINE++G +++VSHD RLI Sbjct: 710 KARVVFAELACREPDVLILDEPTNNLDIESIDALGEAINEYKGAVIVVSHDARLITETNC 769 Query: 1397 EIWVCDHGAVTKWPGDIYSYKTSLIKEI 1480 ++WV + +V++ GD YK +++ + Sbjct: 770 QLWVVEEQSVSQIDGDFDDYKREVLEAL 797
Score = 69.3 bits (168), Expect = 3e-11
Identities = 63/250 (25%), Positives = 108/250 (43%), Gaps = 36/250 (14%)
Frame = +2
Query: 860 FKYGANKPLIYDNIDFGVDLDTRVALVGPNGAGKSTLLK--------------------- 976
F A+ ++ N D + R LVGPNG GK+TLLK
Sbjct: 271 FSISAHGKELFVNADLYIVAGRRYGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQE 330
Query: 977 LIASELEPTDGLIRRHSK-CRIGRYHQHLHEILDL-NSTALEWMMKSFPHIK------EK 1132
++A E ++R +K ++ + L L+ + TA + + K + ++ +
Sbjct: 331 VVADETPAVQAVLRADTKRLKLLEEERRLQGQLEQGDDTAADRLEKVYEELRATGAAAAE 390
Query: 1133 DEMRKKLGRYGLTGGQQICPIKSLSDGQRCRIIFAWLAEGCPHMLLLDEPTNHLDLETID 1312
+ R+ L G Q P + S G R R+ A P +L+LDEPTNHLDL +
Sbjct: 391 AKARRILAGLGFDPEMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVI 450
Query: 1313 SLADAINEFEGGLLLVSHDFRLIGRVAEEIWVCDHGAVTKWPGDIYSYK-------TSLI 1471
L + + + LL+VSHD + V +I D + + G+ ++K L+
Sbjct: 451 WLNNYLQGWRKTLLIVSHDQGFLDDVCTDIIHLDAQRLHYYRGNYMTFKKMYQQKQKELL 510
Query: 1472 KEIEREHTRM 1501
K+ E++ ++
Sbjct: 511 KQYEKQEKKL 520
>sp|Q8NE71|ABCF1_HUMAN ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein) Length = 845 Score = 355 bits (910), Expect = 3e-97 Identities = 197/508 (38%), Positives = 308/508 (60%), Gaps = 16/508 (3%) Frame = +2 Query: 5 NGCGKSTLFAVLSNNELPIPSHIDQFLLQREMPPSDKTALQAVMEADVEKIRLEKEAEYL 184 NG GK+TL ++N L IP +ID L ++E+ + A+QAV+ AD ++++L +E L Sbjct: 338 NGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQAVLRADTKRLKLLEEERRL 397 Query: 185 ALQ----DDTESQDRLIEVYERLXXXXXXXXXXXXXXXXFGLGFTKDMQNKEVKHFSGGW 352 Q DDT ++ RL +VYE L GLGF +MQN+ + FSGGW Sbjct: 398 QGQLEQGDDTAAE-RLEKVYEELRATGAAAAEAKARRILAGLGFDPEMQNRPTQKFSGGW 456 Query: 353 RMRISLARALFVKPALLMLDEPTNHLDLNACVWLEKELSQYNRILLIISHSEDFLNGVCT 532 RMR+SLARALF++P LLMLDEPTNHLDLNA +WL L + + LLI+SH + FL+ VCT Sbjct: 457 RMRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWRKTLLIVSHDQGFLDDVCT 516 Query: 533 NIVHMQNKKLVYYGGNYDQYIMTKMELEENQMKKYKWEQDQMAHMKDYIARFGHGSRKLA 712 +I+H+ ++L YY GNY + + ++ +K+Y+ ++ ++ +K G +++ Sbjct: 517 DIIHLDAQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKLKELK----AGGKSTKQAE 572 Query: 713 RQ-----AQSKEKTLKKMVDGGLTQ-----KVIREKNLVFSFPSCGEIPPPVIQVQNVSF 862 +Q + ++K +K D + K +E + F+FP + PPV+ + V+F Sbjct: 573 KQTKEALTRKQQKCRRKNQDEESQEAPELLKRPKEYTVRFTFPDPPPLSPPVLGLHGVTF 632 Query: 863 KYGANKPLIYDNIDFGVDLDTRVALVGPNGAGKSTLLKLIASELEPTDGLIRRHSKCRIG 1042 Y KPL + N+DFG+D+D+R+ +VGPNG GKSTLL L+ +L PT G +R++ + +IG Sbjct: 633 GYQGQKPL-FKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLTPTHGEMRKNHRLKIG 691 Query: 1043 RYHQHLHEILDLNSTALEWMMKSF--PHIKEKDEMRKKLGRYGLTGGQQICPIKSLSDGQ 1216 ++Q E L + T E++ + F P+ + RK LGR+GL I LS GQ Sbjct: 692 FFNQQYAEQLRMEETPTEYLQRGFNLPY----QDARKCLGRFGLESHAHTIQICKLSGGQ 747 Query: 1217 RCRIIFAWLAEGCPHMLLLDEPTNHLDLETIDSLADAINEFEGGLLLVSHDFRLIGRVAE 1396 + R++FA LA P +L+LDEPTN+LD+E+ID+L +AINE++G +++VSHD RLI Sbjct: 748 KARVVFAELACREPDVLILDEPTNNLDIESIDALGEAINEYKGAVIVVSHDARLITETNC 807 Query: 1397 EIWVCDHGAVTKWPGDIYSYKTSLIKEI 1480 ++WV + +V++ GD YK +++ + Sbjct: 808 QLWVVEEQSVSQIDGDFEDYKREVLEAL 835
Score = 70.5 bits (171), Expect = 1e-11
Identities = 64/250 (25%), Positives = 108/250 (43%), Gaps = 36/250 (14%)
Frame = +2
Query: 860 FKYGANKPLIYDNIDFGVDLDTRVALVGPNGAGKSTLLK--------------------- 976
F A+ ++ N D + R LVGPNG GK+TLLK
Sbjct: 309 FSISAHGKELFVNADLYIVAGRRYGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQE 368
Query: 977 LIASELEPTDGLIRRHSK-CRIGRYHQHLHEILDL-NSTALEWMMKSFPHIK------EK 1132
++A E ++R +K ++ + L L+ + TA E + K + ++ +
Sbjct: 369 VVADETPAVQAVLRADTKRLKLLEEERRLQGQLEQGDDTAAERLEKVYEELRATGAAAAE 428
Query: 1133 DEMRKKLGRYGLTGGQQICPIKSLSDGQRCRIIFAWLAEGCPHMLLLDEPTNHLDLETID 1312
+ R+ L G Q P + S G R R+ A P +L+LDEPTNHLDL +
Sbjct: 429 AKARRILAGLGFDPEMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVI 488
Query: 1313 SLADAINEFEGGLLLVSHDFRLIGRVAEEIWVCDHGAVTKWPGDIYSYK-------TSLI 1471
L + + + LL+VSHD + V +I D + + G+ ++K L+
Sbjct: 489 WLNNYLQGWRKTLLIVSHDQGFLDDVCTDIIHLDAQRLHYYRGNYMTFKKMYQQKQKELL 548
Query: 1472 KEIEREHTRM 1501
K+ E++ ++
Sbjct: 549 KQYEKQEKKL 558
>sp|Q7YR37|ABCF1_PANTR ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) Length = 807 Score = 355 bits (910), Expect = 3e-97 Identities = 197/508 (38%), Positives = 308/508 (60%), Gaps = 16/508 (3%) Frame = +2 Query: 5 NGCGKSTLFAVLSNNELPIPSHIDQFLLQREMPPSDKTALQAVMEADVEKIRLEKEAEYL 184 NG GK+TL ++N L IP +ID L ++E+ + A+QAV+ AD ++++L +E L Sbjct: 300 NGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQAVLRADTKRLKLLEEERRL 359 Query: 185 ALQ----DDTESQDRLIEVYERLXXXXXXXXXXXXXXXXFGLGFTKDMQNKEVKHFSGGW 352 Q DDT ++ RL +VYE L GLGF +MQN+ + FSGGW Sbjct: 360 QGQLEQGDDTAAE-RLEKVYEELRATGAAAAEAKARRILAGLGFDPEMQNRPTQKFSGGW 418 Query: 353 RMRISLARALFVKPALLMLDEPTNHLDLNACVWLEKELSQYNRILLIISHSEDFLNGVCT 532 RMR+SLARALF++P LLMLDEPTNHLDLNA +WL L + + LLI+SH + FL+ VCT Sbjct: 419 RMRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWRKTLLIVSHDQGFLDDVCT 478 Query: 533 NIVHMQNKKLVYYGGNYDQYIMTKMELEENQMKKYKWEQDQMAHMKDYIARFGHGSRKLA 712 +I+H+ ++L YY GNY + + ++ +K+Y+ ++ ++ +K G +++ Sbjct: 479 DIIHLDAQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKLKELK----AGGKSTKQAE 534 Query: 713 RQ-----AQSKEKTLKKMVDGGLTQ-----KVIREKNLVFSFPSCGEIPPPVIQVQNVSF 862 +Q + ++K +K D + K +E + F+FP + PPV+ + V+F Sbjct: 535 KQTKEALTRKQQKCRRKNQDEESQEAPELLKRPKEYTVRFTFPDPPPLSPPVLGLHGVTF 594 Query: 863 KYGANKPLIYDNIDFGVDLDTRVALVGPNGAGKSTLLKLIASELEPTDGLIRRHSKCRIG 1042 Y KPL + N+DFG+D+D+R+ +VGPNG GKSTLL L+ +L PT G +R++ + +IG Sbjct: 595 GYEGQKPL-FKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLTPTHGEMRKNHRLKIG 653 Query: 1043 RYHQHLHEILDLNSTALEWMMKSF--PHIKEKDEMRKKLGRYGLTGGQQICPIKSLSDGQ 1216 ++Q E L + T E++ + F P+ + RK LGR+GL I LS GQ Sbjct: 654 FFNQQYAEQLRMEETPTEYLQRGFNLPY----QDARKCLGRFGLESHAHTIQICKLSGGQ 709 Query: 1217 RCRIIFAWLAEGCPHMLLLDEPTNHLDLETIDSLADAINEFEGGLLLVSHDFRLIGRVAE 1396 + R++FA LA P +L+LDEPTN+LD+E+ID+L +AINE++G +++VSHD RLI Sbjct: 710 KARVVFAELACREPDVLILDEPTNNLDIESIDALGEAINEYKGAVIVVSHDARLITETNC 769 Query: 1397 EIWVCDHGAVTKWPGDIYSYKTSLIKEI 1480 ++WV + +V++ GD YK +++ + Sbjct: 770 QLWVVEEQSVSQIDGDFEDYKREVLEAL 797
Score = 70.9 bits (172), Expect = 1e-11
Identities = 64/250 (25%), Positives = 108/250 (43%), Gaps = 36/250 (14%)
Frame = +2
Query: 860 FKYGANKPLIYDNIDFGVDLDTRVALVGPNGAGKSTLLK--------------------- 976
F A+ ++ N D + R LVGPNG GK+TLLK
Sbjct: 271 FSISAHGKELFVNADLYIVASRRYGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQE 330
Query: 977 LIASELEPTDGLIRRHSK-CRIGRYHQHLHEILDL-NSTALEWMMKSFPHIK------EK 1132
++A E ++R +K ++ + L L+ + TA E + K + ++ +
Sbjct: 331 VVADETPAVQAVLRADTKRLKLLEEERRLQGQLEQGDDTAAERLEKVYEELRATGAAAAE 390
Query: 1133 DEMRKKLGRYGLTGGQQICPIKSLSDGQRCRIIFAWLAEGCPHMLLLDEPTNHLDLETID 1312
+ R+ L G Q P + S G R R+ A P +L+LDEPTNHLDL +
Sbjct: 391 AKARRILAGLGFDPEMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVI 450
Query: 1313 SLADAINEFEGGLLLVSHDFRLIGRVAEEIWVCDHGAVTKWPGDIYSYK-------TSLI 1471
L + + + LL+VSHD + V +I D + + G+ ++K L+
Sbjct: 451 WLNNYLQGWRKTLLIVSHDQGFLDDVCTDIIHLDAQRLHYYRGNYMTFKKMYQQKQKELL 510
Query: 1472 KEIEREHTRM 1501
K+ E++ ++
Sbjct: 511 KQYEKQEKKL 520
>sp|P44808|Y658_HAEIN Probable ABC transporter ATP-binding protein HI0658 Length = 638 Score = 288 bits (738), Expect = 2e-77 Identities = 169/500 (33%), Positives = 279/500 (55%), Gaps = 8/500 (1%) Frame = +2 Query: 2 QNGCGKSTLFAVLSNNELP------IPSHIDQFLLQREMPPSDKTALQAVMEADVEKIRL 163 +NGCGKS+LFA+L +P P++ + +E P D +A+ V++ D E RL Sbjct: 35 KNGCGKSSLFALLKKELMPEGGEVNYPANWRVSWVNQETPALDISAIDYVIQGDREYCRL 94 Query: 164 EKEAEYLALQDDTESQDRLIEVYERLXXXXXXXXXXXXXXXXFGLGFTKDMQNKEVKHFS 343 +++ E ++D + + ++ +L GLGF+++ + VK FS Sbjct: 95 QQKLERANERNDGNA---IARIHGQLETLDAWTIQSRAASLLHGLGFSQEETIQPVKAFS 151 Query: 344 GGWRMRISLARALFVKPALLMLDEPTNHLDLNACVWLEKELSQYNRILLIISHSEDFLNG 523 GGWRMR++LA+AL LL+LDEPTNHLDL+A +WLE+ L QY L++ISH DFL+ Sbjct: 152 GGWRMRLNLAQALLCPSDLLLLDEPTNHLDLDAVIWLERWLVQYQGTLVLISHDRDFLDP 211 Query: 524 VCTNIVHMQNKKLVYYGGNYDQYIMTKMELEENQMKKYKWEQDQMAHMKDYIARFGHGSR 703 + T I+H++N+KL Y G+Y + + + Q Y+ +Q +++H++ YI RF + Sbjct: 212 IVTKILHIENQKLNEYTGDYSSFEVQRATKLAQQTAMYRQQQQKISHLQKYIDRFKAKAT 271 Query: 704 KLARQAQSKEKTLKKMVDGGLTQKVIREKNLVFSFPSCGEIPPPVIQVQNVSFKYGANKP 883 K A+QAQS+ K L++M L + F F +P P++ ++ S YG + Sbjct: 272 K-AKQAQSRMKALERM---ELIAPAYVDNPFTFEFRPPQSLPNPLVMIEQASAGYGIGES 327 Query: 884 L--IYDNIDFGVDLDTRVALVGPNGAGKSTLLKLIASELEPTDGLIRRHSKCRIGRYHQH 1057 I I + +R+ L+G NGAGKSTL+KL+A EL G ++ ++G + QH Sbjct: 328 AVEILSKIKLNLVPGSRIGLLGKNGAGKSTLIKLLAGELTALSGTVQLAKGVQLGYFAQH 387 Query: 1058 LHEILDLNSTALEWMMKSFPHIKEKDEMRKKLGRYGLTGGQQICPIKSLSDGQRCRIIFA 1237 + L + +AL W M+ + + ++R LG + G + +KS S G++ R++ A Sbjct: 388 QLDTLRADESAL-WHMQKLAPEQTEQQVRDYLGSFAFHGDKVNQAVKSFSGGEKARLVLA 446 Query: 1238 WLAEGCPHMLLLDEPTNHLDLETIDSLADAINEFEGGLLLVSHDFRLIGRVAEEIWVCDH 1417 + P++LLLDEPTNHLDL+ +L +A+ ++EG L++VSHD L+ EE ++ Sbjct: 447 LIVWQRPNLLLLDEPTNHLDLDMRQALTEALVDYEGSLVVVSHDRHLLRNTVEEFYLVHD 506 Query: 1418 GAVTKWPGDIYSYKTSLIKE 1477 V ++ GD+ Y+ L ++ Sbjct: 507 KKVEEFKGDLEDYQKWLSEQ 526
Score = 78.2 bits (191), Expect = 6e-14
Identities = 65/251 (25%), Positives = 119/251 (47%), Gaps = 33/251 (13%)
Frame = +2
Query: 836 VIQVQNVSFKYGANKPLIYDNIDFGVDLDTRVALVGPNGAGKSTLLKLIASELEPTDGLI 1015
+I N+S K G + L +N ++ +V LVG NG GKS+L L+ EL P G +
Sbjct: 1 MIIFSNLSLKRGQTELL--ENASATINPKQKVGLVGKNGCGKSSLFALLKKELMPEGGEV 58
Query: 1016 RRHSKCRIGRYHQHLHEILDLNSTALEWMM---KSFPHIKEKDEMRKK------------ 1150
+ R+ +Q E L+ +A+++++ + + +++K E +
Sbjct: 59 NYPANWRVSWVNQ---ETPALDISAIDYVIQGDREYCRLQQKLERANERNDGNAIARIHG 115
Query: 1151 -----------------LGRYGLTGGQQICPIKSLSDGQRCRIIFAWLAEGCPH-MLLLD 1276
L G + + I P+K+ S G R R+ A A CP +LLLD
Sbjct: 116 QLETLDAWTIQSRAASLLHGLGFSQEETIQPVKAFSGGWRMRLNLAQ-ALLCPSDLLLLD 174
Query: 1277 EPTNHLDLETIDSLADAINEFEGGLLLVSHDFRLIGRVAEEIWVCDHGAVTKWPGDIYSY 1456
EPTNHLDL+ + L + +++G L+L+SHD + + +I ++ + ++ GD S+
Sbjct: 175 EPTNHLDLDAVIWLERWLVQYQGTLVLISHDRDFLDPIVTKILHIENQKLNEYTGDYSSF 234
Query: 1457 KTSLIKEIERE 1489
+ ++ ++
Sbjct: 235 EVQRATKLAQQ 245
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 172,513,793
Number of Sequences: 369166
Number of extensions: 3531138
Number of successful extensions: 16530
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 12395
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 15870
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 19268176095
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail