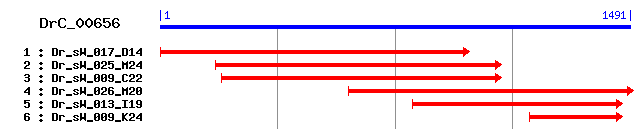
DrC_00656
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00656 (1490 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9DGM7|MVP_ICTPU Major vault protein (MVP) 553 e-157 sp|Q14764|MVP_HUMAN Major vault protein (MVP) (Lung resista... 532 e-151 sp|Q9EQK5|MVP_MOUSE Major vault protein (MVP) 520 e-147 sp|Q62667|MVP_RAT Major vault protein (MVP) 520 e-147 sp|Q90405|MVP_DISOM Major vault protein (MVP100) (P100) 520 e-147 sp|P34118|MVPA_DICDI Major vault protein alpha (MVP-alpha) 509 e-144 sp|P54659|MVPB_DICDI Major vault protein beta (MVP-beta) 427 e-119 sp|P19934|TOLA_ECOLI Protein tolA 59 3e-08 sp|P30427|PLEC1_RAT Plectin 1 (PLTN) (PCN) 52 4e-06 sp|Q9JI55|PLEC1_CRIGR Plectin 1 (PLTN) (PCN) (300-kDa inter... 52 4e-06
>sp|Q9DGM7|MVP_ICTPU Major vault protein (MVP) Length = 871 Score = 553 bits (1425), Expect = e-157 Identities = 285/440 (64%), Positives = 343/440 (77%) Frame = +1 Query: 4 VRSVTGHTYMLTENEELWEKILPDTVEDLLSSCVDPLADRSERNEKPXXXXXXXXXXXXX 183 VR+V GHTYMLT +EELWEK LP VE LLS+ DPLADRSE N+ Sbjct: 426 VRAVIGHTYMLTHDEELWEKELPPNVEKLLSTVRDPLADRSEHNQNAVAEPRDKTMVVS- 484 Query: 184 YRVPHNAAVQIYDYKEKTARIHYGPDLVMLGPDEHFTQLSLSGGKPKKPNLIKSLCLLLG 363 YRVPHNAAVQ+YDY+EK AR+ +GPDLVMLGPDE FT LSLSG KPK+ N+IK++CLLLG Sbjct: 485 YRVPHNAAVQVYDYREKKARVVFGPDLVMLGPDEQFTVLSLSGEKPKRANVIKAICLLLG 544 Query: 364 PDFCTDIIVVETSDHARLSLQVSYNWHFKTEAEKTQNTGTKLFSVPDFVGDACKALASRI 543 PDFCTDII +ET+DHARL LQ++YNWHF+ + LFSVPDFVGDACKA+ASRI Sbjct: 545 PDFCTDIITIETADHARLQLQLAYNWHFEVKNHSDPAQAAALFSVPDFVGDACKAIASRI 604 Query: 544 RGTVAAVRFDDFHKNSARIIRSSVFGVDANNKVREEFVFPQNNLCITSIDIQSVEPVDQR 723 RG VA+V+FDDFHKNS RII S+VFG D VR F QN L I+S+DIQSVEPVDQR Sbjct: 605 RGAVASVQFDDFHKNSIRIICSAVFGFDEKLAVRSSLRFNQNGLVISSVDIQSVEPVDQR 664 Query: 724 TRESLQKSVQLAIEITTKSQEASARHEADRIEQQAKGRLERQMITDQAEAERTRTELLQL 903 TR++LQKSVQLAIEITT SQEA+ARHEA+R+EQ+A+G+LERQ ITDQAEAER R ELL+L Sbjct: 665 TRDALQKSVQLAIEITTNSQEATARHEAERLEQEARGKLERQKITDQAEAERARKELLEL 724 Query: 904 RAASAAVESTGQAKAEAQSRAEASRIEGEANVEQAKLKAEATTIEAENELARVIAARNAE 1083 A SAAVESTG AKAEAQSRAEA+RI+GEA V++AKLK EA IEAE E+ R+ AR E Sbjct: 725 EALSAAVESTGAAKAEAQSRAEAARIQGEAAVQEAKLKVEALRIEAEAEMVRLAKAREQE 784 Query: 1084 IEYSQKHNEVAETHTQKMADIEIYKFSKMVEAIGPNTLIEIAKAGPDGDVRLLQSMGLKS 1263 + Y ++ + + QK+ IE +F ++VEAIG TL +A+AGP+ V++LQ++GLKS Sbjct: 785 LLYKKELDHLEVEKQQKLVTIESQRFKQLVEAIGSETLTAMARAGPELQVKMLQALGLKS 844 Query: 1264 ALITDGSTPINLFQTAHGLI 1323 LITDGS+PINLF TA+GL+ Sbjct: 845 TLITDGSSPINLFTTANGLL 864
>sp|Q14764|MVP_HUMAN Major vault protein (MVP) (Lung resistance-related protein) Length = 893 Score = 532 bits (1371), Expect = e-151 Identities = 272/452 (60%), Positives = 345/452 (76%), Gaps = 12/452 (2%) Frame = +1 Query: 4 VRSVTGHTYMLTENEELWEKILPDTVEDLLSSCVDPLADRSERN--EKPXXXXXXXXXXX 177 VR+V G TYMLT++E LWEK LP VE+LL+ DPLADR E++ + Sbjct: 398 VRAVIGSTYMLTQDEVLWEKELPPGVEELLNKGQDPLADRGEKDTAKSLQPLAPRNKTRV 457 Query: 178 XXYRVPHNAAVQIYDYKEKTARIHYGPDLVMLGPDEHFTQLSLSGGKPKKPNLIKSLCLL 357 YRVPHNAAVQ+YDY+EK AR+ +GP+LV LGP+E FT LSLS G+PK+P+ ++LCLL Sbjct: 458 VSYRVPHNAAVQVYDYREKRARVVFGPELVSLGPEEQFTVLSLSAGRPKRPHARRALCLL 517 Query: 358 LGPDFCTDIIVVETSDHARLSLQVSYNWHFKTEAEKTQNTGTKLFSVPDFVGDACKALAS 537 LGPDF TD+I +ET+DHARL LQ++YNWHF+ K KLFSVPDFVGDACKA+AS Sbjct: 518 LGPDFFTDVITIETADHARLQLQLAYNWHFEVNDRKDPQETAKLFSVPDFVGDACKAIAS 577 Query: 538 RIRGTVAAVRFDDFHKNSARIIRSSVFGVDANN----------KVREEFVFPQNNLCITS 687 R+RG VA+V FDDFHKNSARIIR++VFG + + + R++ VFPQN L ++S Sbjct: 578 RVRGAVASVTFDDFHKNSARIIRTAVFGFETSEAKGPDGMALPRPRDQAVFPQNGLVVSS 637 Query: 688 IDIQSVEPVDQRTRESLQKSVQLAIEITTKSQEASARHEADRIEQQAKGRLERQMITDQA 867 +D+QSVEPVDQRTR++LQ+SVQLAIEITT SQEA+A+HEA R+EQ+A+GRLERQ I DQ+ Sbjct: 638 VDVQSVEPVDQRTRDALQRSVQLAIEITTNSQEAAAKHEAQRLEQEARGRLERQKILDQS 697 Query: 868 EAERTRTELLQLRAASAAVESTGQAKAEAQSRAEASRIEGEANVEQAKLKAEATTIEAEN 1047 EAE+ R ELL+L A S AVESTG AKAEA+SRAEA+RIEGE +V QAKLKA+A IE E Sbjct: 698 EAEKARKELLELEALSMAVESTGTAKAEAESRAEAARIEGEGSVLQAKLKAQALAIETEA 757 Query: 1048 ELARVIAARNAEIEYSQKHNEVAETHTQKMADIEIYKFSKMVEAIGPNTLIEIAKAGPDG 1227 EL RV R E+ Y++ E+ + Q++A++E+ KF +M EAIGP+T+ ++A AGP+ Sbjct: 758 ELQRVQKVRELELVYARAQLELEVSKAQQLAEVEVKKFKQMTEAIGPSTIRDLAVAGPEM 817 Query: 1228 DVRLLQSMGLKSALITDGSTPINLFQTAHGLI 1323 V+LLQS+GLKS LITDGSTPINLF TA GL+ Sbjct: 818 QVKLLQSLGLKSTLITDGSTPINLFNTAFGLL 849
>sp|Q9EQK5|MVP_MOUSE Major vault protein (MVP) Length = 861 Score = 520 bits (1339), Expect = e-147 Identities = 268/452 (59%), Positives = 337/452 (74%), Gaps = 12/452 (2%) Frame = +1 Query: 4 VRSVTGHTYMLTENEELWEKILPDTVEDLLSSCVDPLADRSERNEKPXXXXXXXXXXXXX 183 VR+V G TYMLT++E LWEK LP VE+LL+ DPLADR ++ Sbjct: 398 VRAVIGSTYMLTQDEVLWEKELPSGVEELLNLGHDPLADRGQKGTAKVLQPSAARNKTRV 457 Query: 184 --YRVPHNAAVQIYDYKEKTARIHYGPDLVMLGPDEHFTQLSLSGGKPKKPNLIKSLCLL 357 YRVPHNAAVQ+YDY+ K AR+ +GP+LV L P+E FT LSLS G+PK+P+ ++LCLL Sbjct: 458 VSYRVPHNAAVQVYDYRAKRARVVFGPELVSLDPEEQFTVLSLSAGRPKRPHARRALCLL 517 Query: 358 LGPDFCTDIIVVETSDHARLSLQVSYNWHFKTEAEKTQNTGTKLFSVPDFVGDACKALAS 537 LGPDF TD+I +ET+DHARL LQ++YNWHF+ + KLFSVPDFVGDACKA+AS Sbjct: 518 LGPDFFTDVITIETADHARLQLQLAYNWHFELKNRNDPEETAKLFSVPDFVGDACKAIAS 577 Query: 538 RIRGTVAAVRFDDFHKNSARIIRSSVFGVDANN----------KVREEFVFPQNNLCITS 687 R+RG VA+V FDDFHKNSARIIR +VFG + + + R+ VFPQN L ++S Sbjct: 578 RVRGAVASVTFDDFHKNSARIIRMAVFGFEMSEDAGPDGALLPRARDRAVFPQNGLVVSS 637 Query: 688 IDIQSVEPVDQRTRESLQKSVQLAIEITTKSQEASARHEADRIEQQAKGRLERQMITDQA 867 +D+QSVEPVDQRTR++LQ+SVQLAIEITT SQEA+A+HEA R+EQ+A+GRLERQ I DQ+ Sbjct: 638 VDVQSVEPVDQRTRDALQRSVQLAIEITTNSQEAAAKHEAQRLEQEARGRLERQKILDQS 697 Query: 868 EAERTRTELLQLRAASAAVESTGQAKAEAQSRAEASRIEGEANVEQAKLKAEATTIEAEN 1047 EAE+ R ELL+L A S AVESTG AKAEA+SRAEA+RIEGE +V QAKLKA+A IE E Sbjct: 698 EAEKARKELLELEAMSMAVESTGNAKAEAESRAEAARIEGEGSVLQAKLKAQALAIETEA 757 Query: 1048 ELARVIAARNAEIEYSQKHNEVAETHTQKMADIEIYKFSKMVEAIGPNTLIEIAKAGPDG 1227 EL RV R E+ YS+ E+ + Q++AD+E KF +M EA+GP T+ ++A AGP+ Sbjct: 758 ELERVKKVREMELIYSRAQLELEVSKAQQLADVEAKKFKEMTEALGPGTIRDLAVAGPEM 817 Query: 1228 DVRLLQSMGLKSALITDGSTPINLFQTAHGLI 1323 V+LLQS+GLKS LITDGS+PINLF TA GL+ Sbjct: 818 QVKLLQSLGLKSTLITDGSSPINLFNTAFGLL 849
>sp|Q62667|MVP_RAT Major vault protein (MVP) Length = 861 Score = 520 bits (1339), Expect = e-147 Identities = 269/452 (59%), Positives = 340/452 (75%), Gaps = 12/452 (2%) Frame = +1 Query: 4 VRSVTGHTYMLTENEELWEKILPDTVEDLLSSCVDPLADRSERNE-KPXXXXXXXXXXXX 180 VR+V G TYMLT++E LWEK LP VE+LL+ DPLADR ++ KP Sbjct: 398 VRAVIGSTYMLTQDEVLWEKELPSGVEELLNLGHDPLADRGQKGTAKPLQPSAPRNKTRV 457 Query: 181 X-YRVPHNAAVQIYDYKEKTARIHYGPDLVMLGPDEHFTQLSLSGGKPKKPNLIKSLCLL 357 YRVPHNAAVQ+YDY+ K AR+ +GP+LV L P+E FT LSLS G+PK+P+ ++LCLL Sbjct: 458 VSYRVPHNAAVQVYDYRAKRARVVFGPELVTLDPEEQFTVLSLSAGRPKRPHARRALCLL 517 Query: 358 LGPDFCTDIIVVETSDHARLSLQVSYNWHFKTEAEKTQNTGTKLFSVPDFVGDACKALAS 537 LGPDF TD+I +ET+DHARL LQ++YNWHF+ + KLFSVPDFVGDACKA+AS Sbjct: 518 LGPDFFTDVITIETADHARLQLQLAYNWHFELKNRNDPAEAAKLFSVPDFVGDACKAIAS 577 Query: 538 RIRGTVAAVRFDDFHKNSARIIRSSVFGVDANN----------KVREEFVFPQNNLCITS 687 R+RG VA+V FDDFHKNSARIIR +VFG + + K R++ VFPQN L ++S Sbjct: 578 RVRGAVASVTFDDFHKNSARIIRMAVFGFEMSEDTGPDGTLLPKARDQAVFPQNGLVVSS 637 Query: 688 IDIQSVEPVDQRTRESLQKSVQLAIEITTKSQEASARHEADRIEQQAKGRLERQMITDQA 867 +D+QSVEPVDQRTR++LQ+SVQLAIEITT SQEA+A+HEA R+EQ+A+GRLERQ I DQ+ Sbjct: 638 VDVQSVEPVDQRTRDALQRSVQLAIEITTNSQEAAAKHEAQRLEQEARGRLERQKILDQS 697 Query: 868 EAERTRTELLQLRAASAAVESTGQAKAEAQSRAEASRIEGEANVEQAKLKAEATTIEAEN 1047 EAE+ R ELL+L A S AVESTG AKAEA+SRAEA+RIEGE +V QAKLKA+A IE E Sbjct: 698 EAEKARKELLELEAMSMAVESTGNAKAEAESRAEAARIEGEGSVLQAKLKAQALAIETEA 757 Query: 1048 ELARVIAARNAEIEYSQKHNEVAETHTQKMADIEIYKFSKMVEAIGPNTLIEIAKAGPDG 1227 EL RV R E+ Y++ E+ + Q++A++E KF +M EA+GP T+ ++A AGP+ Sbjct: 758 ELERVKKVREMELIYARAQLELEVSKAQQLANVEAKKFKEMTEALGPGTIRDLAVAGPEM 817 Query: 1228 DVRLLQSMGLKSALITDGSTPINLFQTAHGLI 1323 V+LLQS+GLKS LITDGS+PINLF TA GL+ Sbjct: 818 QVKLLQSLGLKSTLITDGSSPINLFSTAFGLL 849
>sp|Q90405|MVP_DISOM Major vault protein (MVP100) (P100) Length = 852 Score = 520 bits (1339), Expect = e-147 Identities = 263/444 (59%), Positives = 340/444 (76%), Gaps = 4/444 (0%) Frame = +1 Query: 4 VRSVTGHTYMLTENEELWEKILPDTVEDLLSSCVDPLADR----SERNEKPXXXXXXXXX 171 VR+V G +YML+++EELWEK LP VE LL+ DP+ +R SE + Sbjct: 396 VRAVIGESYMLSQDEELWEKTLPPNVEALLTPDKDPVLNRNVHQSEAGHQEEGVKKRDPT 455 Query: 172 XXXXYRVPHNAAVQIYDYKEKTARIHYGPDLVMLGPDEHFTQLSLSGGKPKKPNLIKSLC 351 +RVPHNAAVQ+YDY +K+AR+ +GP+LVML DE T LSLSGG+PK+P +I+SLC Sbjct: 456 RVVTFRVPHNAAVQVYDYSQKSARVVFGPELVMLETDEQLTVLSLSGGRPKRPGVIRSLC 515 Query: 352 LLLGPDFCTDIIVVETSDHARLSLQVSYNWHFKTEAEKTQNTGTKLFSVPDFVGDACKAL 531 LLLGPDF TD+IV+ET+DHARL LQ++YNWHF+ + K+FSV DFVGDACKA+ Sbjct: 516 LLLGPDFFTDVIVIETADHARLQLQLAYNWHFEVKDRSDPMESAKVFSVLDFVGDACKAI 575 Query: 532 ASRIRGTVAAVRFDDFHKNSARIIRSSVFGVDANNKVREEFVFPQNNLCITSIDIQSVEP 711 ASRIRG VA+V+FDDFHKNS+RI+R++VFG+D +VRE +FPQN+LC+TS+D+ SVEP Sbjct: 576 ASRIRGAVASVQFDDFHKNSSRILRAAVFGLDPELRVRERLLFPQNSLCVTSVDVHSVEP 635 Query: 712 VDQRTRESLQKSVQLAIEITTKSQEASARHEADRIEQQAKGRLERQMITDQAEAERTRTE 891 VDQRTR++LQ+SVQLAIEITTKSQEA+AR EA R+EQ+AKGRLERQ I DQAE+ER R E Sbjct: 636 VDQRTRDALQRSVQLAIEITTKSQEAAARQEAARLEQEAKGRLERQRILDQAESERARRE 695 Query: 892 LLQLRAASAAVESTGQAKAEAQSRAEASRIEGEANVEQAKLKAEATTIEAENELARVIAA 1071 LL L A S AVES G A+AEA+SRAEASRIEGE +V QAKLKA+A IE E EL R+ A Sbjct: 696 LLLLEADSTAVESRGAARAEAESRAEASRIEGEGSVLQAKLKAQALDIETEAELERLTKA 755 Query: 1072 RNAEIEYSQKHNEVAETHTQKMADIEIYKFSKMVEAIGPNTLIEIAKAGPDGDVRLLQSM 1251 R E++Y + N + T++++++E +F +MV+AIG +T+ IA++GP+ V+LLQ + Sbjct: 756 RERELKYIAEQNRLEVEKTRQLSELESRRFQEMVQAIGADTIRAIAESGPELQVKLLQGL 815 Query: 1252 GLKSALITDGSTPINLFQTAHGLI 1323 GL S LITDGSTP+NLF A G++ Sbjct: 816 GLNSTLITDGSTPVNLFSAAKGML 839
>sp|P34118|MVPA_DICDI Major vault protein alpha (MVP-alpha) Length = 843 Score = 509 bits (1312), Expect = e-144 Identities = 263/440 (59%), Positives = 333/440 (75%) Frame = +1 Query: 4 VRSVTGHTYMLTENEELWEKILPDTVEDLLSSCVDPLADRSERNEKPXXXXXXXXXXXXX 183 V S+ G +YML NEELWEK+LP TVE++L+ + + P Sbjct: 395 VASIKGQSYMLKANEELWEKVLPRTVEEVLA----------KESLNPEFTDNRDSTRVVT 444 Query: 184 YRVPHNAAVQIYDYKEKTARIHYGPDLVMLGPDEHFTQLSLSGGKPKKPNLIKSLCLLLG 363 YR PHN+AVQIYDYKEK +R+ +GPDLVMLGPDEHFT LSLSG KPK+P+ IK++ L LG Sbjct: 445 YRAPHNSAVQIYDYKEKKSRVVFGPDLVMLGPDEHFTVLSLSGDKPKRPHQIKAIALFLG 504 Query: 364 PDFCTDIIVVETSDHARLSLQVSYNWHFKTEAEKTQNTGTKLFSVPDFVGDACKALASRI 543 PDF TD+++VETSDHARLSL++SYNW FK + K ++ K+F VPDFVGD+CKA+ASR+ Sbjct: 505 PDFMTDVVIVETSDHARLSLKLSYNWEFKVDRSKIEDA-QKIFQVPDFVGDSCKAIASRV 563 Query: 544 RGTVAAVRFDDFHKNSARIIRSSVFGVDANNKVREEFVFPQNNLCITSIDIQSVEPVDQR 723 RG VAAV FDDFHK SA +IR SVFG+D + +VR+ F F NNL IT+IDIQSVEPVDQR Sbjct: 564 RGAVAAVSFDDFHKRSAEVIRQSVFGLDESGEVRKNFSFNSNNLVITNIDIQSVEPVDQR 623 Query: 724 TRESLQKSVQLAIEITTKSQEASARHEADRIEQQAKGRLERQMITDQAEAERTRTELLQL 903 TR+SLQKSVQLAIEITTKSQEA+ARHEA+R+EQ A+GRLERQ I D+A+AE R +LLQL Sbjct: 624 TRDSLQKSVQLAIEITTKSQEAAARHEAERLEQGARGRLERQKIHDEAQAELARKDLLQL 683 Query: 904 RAASAAVESTGQAKAEAQSRAEASRIEGEANVEQAKLKAEATTIEAENELARVIAARNAE 1083 +A SAAVESTGQA AEAQ+ AEA+ I EANV+QA+LKA+AT I +E+E++ + A R E Sbjct: 684 QAQSAAVESTGQATAEAQATAEAANIAAEANVKQAELKAQATKIRSESEISLLKAKRENE 743 Query: 1084 IEYSQKHNEVAETHTQKMADIEIYKFSKMVEAIGPNTLIEIAKAGPDGDVRLLQSMGLKS 1263 + Y + +E+ T +A+IE KF +VE+IG +TL IA AG + +LLQ +GLKS Sbjct: 744 LSYQKSIDELELTKQSDLAEIEASKFKAIVESIGRDTLKSIACAGNEMQAKLLQGLGLKS 803 Query: 1264 ALITDGSTPINLFQTAHGLI 1323 +ITDG +P+NLF TA+G+I Sbjct: 804 FMITDGKSPLNLFDTANGII 823
>sp|P54659|MVPB_DICDI Major vault protein beta (MVP-beta) Length = 846 Score = 427 bits (1098), Expect = e-119 Identities = 235/440 (53%), Positives = 305/440 (69%) Frame = +1 Query: 4 VRSVTGHTYMLTENEELWEKILPDTVEDLLSSCVDPLADRSERNEKPXXXXXXXXXXXXX 183 VRSV G +YML NEE W K L VE+LL D++E + + Sbjct: 407 VRSVHGSSYMLLPNEERWAKPLAPIVEELLKKSAS--RDQNEEHNESDRDSTKVIT---- 460 Query: 184 YRVPHNAAVQIYDYKEKTARIHYGPDLVMLGPDEHFTQLSLSGGKPKKPNLIKSLCLLLG 363 YRVPHNAAVQI+DY++K AR+ +GP+LVML PDE FT LSLSG PK+PN I+SL L LG Sbjct: 461 YRVPHNAAVQIFDYRKKEARVVFGPELVMLEPDEEFTVLSLSGKIPKRPNHIRSLALFLG 520 Query: 364 PDFCTDIIVVETSDHARLSLQVSYNWHFKTEAEKTQNTGTKLFSVPDFVGDACKALASRI 543 PDF TD+I VET+DHARLSL++SYNWHFK E Q+ +KLF+ DF GD CKA S + Sbjct: 521 PDFMTDLITVETADHARLSLKLSYNWHFKVE----QDNPSKLFATSDFTGDLCKATGSLV 576 Query: 544 RGTVAAVRFDDFHKNSARIIRSSVFGVDANNKVREEFVFPQNNLCITSIDIQSVEPVDQR 723 R VAA FD+FHK+S+ II+ +VFG + + F N L IT+ID+QSVEPVDQR Sbjct: 577 RAAVAASTFDNFHKHSSDIIQQAVFG-STDGSSNDCLYFETNGLVITNIDVQSVEPVDQR 635 Query: 724 TRESLQKSVQLAIEITTKSQEASARHEADRIEQQAKGRLERQMITDQAEAERTRTELLQL 903 T +SLQKSVQLAIEITTKSQEA+AR EA+R+EQ A+G LERQ I D+A+ E +R++L+QL Sbjct: 636 TLDSLQKSVQLAIEITTKSQEATARQEAERLEQMARGELERQKIIDEAKNEESRSKLVQL 695 Query: 904 RAASAAVESTGQAKAEAQSRAEASRIEGEANVEQAKLKAEATTIEAENELARVIAARNAE 1083 +A SAAVESTGQA AEA++RAEA+ IE E+ ++QA+L A IEA +E+ + A E Sbjct: 696 QAQSAAVESTGQAVAEARARAEAAIIEAESEIKQARLSTRAIEIEALSEIENLKAKHIVE 755 Query: 1084 IEYSQKHNEVAETHTQKMADIEIYKFSKMVEAIGPNTLIEIAKAGPDGDVRLLQSMGLKS 1263 I++ + N + ++ A IE KF VEA+G +T+ IA+A + +LL +GLKS Sbjct: 756 IQHIKSLNNLELIKAKESATIETTKFENYVEALGSDTIKSIAQAPEETKAKLLAGLGLKS 815 Query: 1264 ALITDGSTPINLFQTAHGLI 1323 +ITDG +P+NLF TA+GLI Sbjct: 816 FMITDGKSPLNLFDTANGLI 835
>sp|P19934|TOLA_ECOLI Protein tolA Length = 421 Score = 59.3 bits (142), Expect = 3e-08 Identities = 48/158 (30%), Positives = 74/158 (46%) Frame = +1 Query: 694 IQSVEPVDQRTRESLQKSVQLAIEITTKSQEASARHEADRIEQQAKGRLERQMITDQAEA 873 +QS E +R+ E + Q A E + Q A E +R++Q K RL Q QAE Sbjct: 67 MQSQESSAKRSDEQRKMKEQQAAEELREKQAA----EQERLKQLEKERLAAQEQKKQAEE 122 Query: 874 ERTRTELLQLRAASAAVESTGQAKAEAQSRAEASRIEGEANVEQAKLKAEATTIEAENEL 1053 + EL Q +A AA ++ AKA+A++ A+A+ + AK KAEA +A E Sbjct: 123 AAKQAELKQKQAEEAAAKAAADAKAKAEADAKAAEEAAKKAAADAKKKAEAEAAKAAAEA 182 Query: 1054 ARVIAARNAEIEYSQKHNEVAETHTQKMADIEIYKFSK 1167 + A A ++ + E A +K A E + +K Sbjct: 183 QKKAEAAAAALKKKAEAAEAAAAEARKKAATEAAEKAK 220
>sp|P30427|PLEC1_RAT Plectin 1 (PLTN) (PCN) Length = 4687 Score = 52.0 bits (123), Expect = 4e-06 Identities = 51/169 (30%), Positives = 79/169 (46%), Gaps = 17/169 (10%) Frame = +1 Query: 694 IQSVEPVDQRTRESLQKSVQLAIEITTKSQEASARHEADRIEQQAKGRLERQMITDQAEA 873 ++ + +R + + +QLA E K +A + A + QQ + L++ + +Q Sbjct: 2158 VEEARRLRERAEQESARQLQLAQEAAQKRLQAEEKAHAF-VVQQREEELQQTLQQEQNML 2216 Query: 874 ERTRTEL-LQLRAASAAVESTGQAKAEA-QSRA---EASRIEGEANVE-----QAKLKAE 1023 ER R+E RAA A E+ QA+ EA QSR EA R++ A + QA+ AE Sbjct: 2217 ERLRSEAEAARRAAEEAEEAREQAEREAAQSRKQVEEAERLKQSAEEQAQAQAQAQAAAE 2276 Query: 1024 ATTIEAENELARVIAARNAEIEYSQ-------KHNEVAETHTQKMADIE 1149 EAE E AR A A ++ Q KH + AE ++ A +E Sbjct: 2277 KLRKEAEQEAARRAQAEQAALKQKQAADAEMEKHKKFAEQTLRQKAQVE 2325
Score = 43.1 bits (100), Expect = 0.002
Identities = 45/167 (26%), Positives = 70/167 (41%), Gaps = 18/167 (10%)
Frame = +1
Query: 712 VDQRTRESLQKSVQLAIEITTKSQEASARHEA----DRIEQQAKGRLERQMITDQAEAER 879
V ++ Q +LA+ + +++ A + A D ++ QA+ E + QAEAER
Sbjct: 1643 VQDESQRKRQAEAELALRVKAEAEAAREKQRALQALDELKLQAE---EAERWLCQAEAER 1699
Query: 880 TRTELLQLRAA--SAAVESTGQAKAEAQSRAEASRIEGEANVEQAKLKAEA--------- 1026
R + L A SA VE + + A+ A+ R E +V +L+ EA
Sbjct: 1700 ARQVQVALETAQRSAEVELQSKRPSFAEKTAQLERTLQEEHVTVTQLREEAERRAQQQAE 1759
Query: 1027 ---TTIEAENELARVIAARNAEIEYSQKHNEVAETHTQKMADIEIYK 1158
EAE EL R N + + EVA+ + AD E K
Sbjct: 1760 AERAREEAERELERWQLKANEALRLRLQAEEVAQQKSLAQADAEKQK 1806
Score = 37.4 bits (85), Expect = 0.11
Identities = 46/175 (26%), Positives = 79/175 (45%), Gaps = 12/175 (6%)
Frame = +1
Query: 721 RTRESLQKSVQLAIEIT--TKSQEASARHEADRIEQQAK-GRLERQMITD-----QAEAE 876
R RE ++ ++ ++T T Q +A E R+ + + G +RQ++ + Q EA
Sbjct: 1825 RQRELAEQELEKQRQLTEGTAQQRLAAEQELIRLRAETEQGEHQRQLLEEELARLQHEAT 1884
Query: 877 RTRTELLQLRAASAAVESTGQAKAEAQSRAEASRIEGEANVEQAKLKAEATTIEAENELA 1056
+ +L A A V + + +++RAE E + E++K + EA ELA
Sbjct: 1885 AATQKRQELEAELAKVRAEMEVLLASKARAEE---ESRSTSEKSKQRLEAEAGRFR-ELA 1940
Query: 1057 RVIAARNAEIEYSQKHNEVAETHTQKM---AD-IEIYKFSKMVEAIGPNTLIEIA 1209
A A E +++H E+AE + AD + K + + EA T EIA
Sbjct: 1941 EEAARLRALAEEARRHRELAEEDAARQRAEADGVLTEKLAAISEATRLKTEAEIA 1995
Score = 36.2 bits (82), Expect = 0.25
Identities = 36/136 (26%), Positives = 62/136 (45%), Gaps = 10/136 (7%)
Frame = +1
Query: 697 QSVEPVDQRTRESLQKSVQLAIE-ITTKSQEASARHEADRIEQQAKGRLERQMITDQAEA 873
Q +E Q T + Q+ + E I +++ H+ +E++ RL+ + +
Sbjct: 1832 QELEKQRQLTEGTAQQRLAAEQELIRLRAETEQGEHQRQLLEEEL-ARLQHEATAATQKR 1890
Query: 874 ERTRTELLQLRAA-SAAVESTGQAKAEAQSRAEAS--RIEGEAN------VEQAKLKAEA 1026
+ EL ++RA + S +A+ E++S +E S R+E EA E A+L+A A
Sbjct: 1891 QELEAELAKVRAEMEVLLASKARAEEESRSTSEKSKQRLEAEAGRFRELAEEAARLRALA 1950
Query: 1027 TTIEAENELARVIAAR 1074
ELA AAR
Sbjct: 1951 EEARRHRELAEEDAAR 1966
Score = 35.0 bits (79), Expect = 0.56
Identities = 42/163 (25%), Positives = 75/163 (46%), Gaps = 2/163 (1%)
Frame = +1
Query: 697 QSVEPVDQRTRESLQKSVQLAIEITTKSQEASARHEADRIEQQAKGRLER-QMITDQAEA 873
+ ++ V + TR L+ +L + +QE + R +AD+ EQ A+ +E Q EA
Sbjct: 2454 EKMQAVQEATR--LKAEAELLQQQKELAQEQARRLQADK-EQMAQQLVEETQGFQRTLEA 2510
Query: 874 ERTRTELLQLRAASAAVESTGQAKAEAQSRAEASRIEGEANVEQAKL-KAEATTIEAENE 1050
ER R + A + ++A+A++ +A R +A KL + E T E
Sbjct: 2511 ERQRQLEMSAEAERLKLRMAEMSRAQARAEEDAQRFRKQAEEIGEKLHRTELATQE---- 2566
Query: 1051 LARVIAARNAEIEYSQKHNEVAETHTQKMADIEIYKFSKMVEA 1179
+V + EI+ Q+ ++ AE + +A++E K EA
Sbjct: 2567 --KVTLVQTLEIQ-RQQSDQDAERLREAIAELEREKEKLKQEA 2606
Score = 31.6 bits (70), Expect = 6.2
Identities = 31/130 (23%), Positives = 56/130 (43%), Gaps = 10/130 (7%)
Frame = +1
Query: 781 QEASARHEAD------RIEQQAKGRLERQ--MITDQAEAERTRTELLQLRAASAAVESTG 936
+E +A+H+AD ++ + ++ LERQ ++ D R E ++ A A+ E
Sbjct: 2021 EEQAAQHKADIEERLAQLRKASESELERQKGLVEDTLRQRRQVEE--EIMALKASFEKAA 2078
Query: 937 QAKAEAQSRAEASRIEGEANVEQAKLKAEATTIEAENELARVIAARNAEI--EYSQKHNE 1110
KAE + +E ++++ A ELA AAR ++ E Q+ E
Sbjct: 2079 AGKAELE-------------LELGRIRSNAEDTMRSKELAEQEAARQRQLAAEEEQRRRE 2125
Query: 1111 VAETHTQKMA 1140
E + +A
Sbjct: 2126 AEERVQRSLA 2135
Score = 31.2 bits (69), Expect = 8.2
Identities = 18/59 (30%), Positives = 29/59 (49%), Gaps = 2/59 (3%)
Frame = +1
Query: 196 HNAAVQIYDYKEKTARIHYGPDLVMLGPDE--HFTQLSLSGGKPKKPNLIKSLCLLLGP 366
H + + DYK+ +H G ++GP + H+ LS SG + P S+C L+ P
Sbjct: 946 HVPLLAVCDYKQVEVTVHKGDQCQLVGPAQPFHWKVLSSSGSEAAVP----SVCFLVPP 1000
>sp|Q9JI55|PLEC1_CRIGR Plectin 1 (PLTN) (PCN) (300-kDa intermediate filament-associated protein) (IFAP300) Length = 4473 Score = 52.0 bits (123), Expect = 4e-06 Identities = 51/169 (30%), Positives = 79/169 (46%), Gaps = 17/169 (10%) Frame = +1 Query: 694 IQSVEPVDQRTRESLQKSVQLAIEITTKSQEASARHEADRIEQQAKGRLERQMITDQAEA 873 ++ + +R + + +QLA E K +A + A + QQ + L++ + +Q+ Sbjct: 1944 VEEARRLRERAEQESARQLQLAQEAAQKRLQAEEKAHAF-VVQQREEELQQTLQQEQSML 2002 Query: 874 ERTRTEL-LQLRAASAAVESTGQAKAEA-QSRA---EASRIEGEANVE-----QAKLKAE 1023 ER R E RAA A E+ QA+ EA QSR EA R++ A + QA+ AE Sbjct: 2003 ERLRGEAEAARRAAEEAEEAREQAEREAAQSRKQVEEAERLKQSAEEQAQARAQAQAAAE 2062 Query: 1024 ATTIEAENELARVIAARNAEIEYSQ-------KHNEVAETHTQKMADIE 1149 EAE E AR A A ++ Q KH + AE ++ A +E Sbjct: 2063 KLRKEAEQEAARRAQAEQAALKQKQAADAEMEKHKKFAEQTLRQKAQVE 2111
Score = 42.0 bits (97), Expect = 0.005
Identities = 42/164 (25%), Positives = 67/164 (40%), Gaps = 15/164 (9%)
Frame = +1
Query: 712 VDQRTRESLQKSVQLAIEITTKSQEASARHEA-DRIEQQAKGRLERQMITDQAEAERTRT 888
V ++ Q +LA+ + +++ A + A +E+ E + QA+AER R
Sbjct: 1429 VQDESQRKRQAEAELALRVKAQAEAAQEKQRALQALEELRLQAEEAERRLRQAQAERARQ 1488
Query: 889 ELLQLRAA--SAAVESTGQAKAEAQSRAEASRIEGEANVEQAKLK------------AEA 1026
+ L A SA VE + + A+ A+ R E +V +L+ AE
Sbjct: 1489 VQVALETAQRSAEVELQSKRASFAEKTAQLERTLQEEHVTVTQLREKAERRAQQQAEAER 1548
Query: 1027 TTIEAENELARVIAARNAEIEYSQKHNEVAETHTQKMADIEIYK 1158
EAE EL R N + + EVA+ + AD E K
Sbjct: 1549 AREEAERELERWQLKANEALRLRLQAEEVAQQRSLAQADAEKQK 1592
Score = 34.3 bits (77), Expect = 0.96
Identities = 33/154 (21%), Positives = 67/154 (43%), Gaps = 4/154 (2%)
Frame = +1
Query: 730 ESLQKSVQLAIEITTKSQEASARHEADRIEQQAKGRLERQMITDQAEAERT----RTELL 897
+++Q++ +L E Q+ E R Q+ K ++ +Q++ + +RT R L
Sbjct: 2243 QAVQEATRLKAEAELLQQQKELAQEQARRLQEDKEQMAQQLVEETQGFQRTLEVERQRQL 2302
Query: 898 QLRAASAAVESTGQAKAEAQSRAEASRIEGEANVEQAKLKAEATTIEAENELARVIAARN 1077
++ A + ++ + AQ+RAE E+ K T + + +V +
Sbjct: 2303 EMSAEAERLKLRMAEMSRAQARAEEDAQRFRKQAEEIGEKLHRTELATQE---KVTLVQT 2359
Query: 1078 AEIEYSQKHNEVAETHTQKMADIEIYKFSKMVEA 1179
EI+ Q ++ AE + +A++E K EA
Sbjct: 2360 LEIQRQQSDHD-AERLREAIAELEREKEKLKQEA 2392
Score = 32.0 bits (71), Expect = 4.8
Identities = 46/187 (24%), Positives = 84/187 (44%), Gaps = 30/187 (16%)
Frame = +1
Query: 679 ITSIDIQSVEPVDQRT--RESLQKSVQLAIEITTKSQEASARHE---ADRIEQQAKGRLE 843
+T++ +Q E Q++ E LQ+ L E+T +++ S E + R++ + G+L+
Sbjct: 2114 LTTLRLQLEETDHQKSILDEELQR---LKAEVTEAARQRSQVEEELFSVRVQMEELGKLK 2170
Query: 844 RQMITD----------------QAEAERTRTELLQLRAASAAVESTGQAK--AE---AQS 960
++ + + EAE+ + + S A + + + AE AQ
Sbjct: 2171 ARIEAENRALILRDKDNTQRFLEEEAEKMKQVAEEAARLSVAAQEAARLRQLAEEDLAQQ 2230
Query: 961 RAEASRIEGE---ANVEQAKLKAEATTIEAENELARVIAARNAEIEYSQKHNEVAETH-T 1128
RA A ++ E A E +LKAEA ++ + ELA+ A R E + V ET
Sbjct: 2231 RALAEKMLKEKMQAVQEATRLKAEAELLQQQKELAQEQARRLQEDKEQMAQQLVEETQGF 2290
Query: 1129 QKMADIE 1149
Q+ ++E
Sbjct: 2291 QRTLEVE 2297
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 151,915,844
Number of Sequences: 369166
Number of extensions: 2978679
Number of successful extensions: 9219
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8638
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9126
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 17949083355
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail