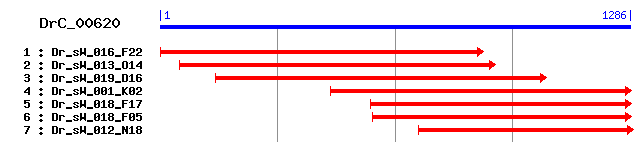
DrC_00620
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00620 (1285 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9VCI3|LSD1_DROME Lipid storage droplets surface binding... 55 4e-07 sp|Q9VXY7|LSD2_DROME Lipid storage droplets surface binding... 49 3e-05 sp|P43883|ADFP_MOUSE Adipophilin (Adipose differentiation-r... 47 1e-04 sp|Q9TUM6|ADFP_BOVIN Adipophilin (Adipose differentiation-r... 44 0.001 sp|Q9N4M4|ANC1_CAEEL Nuclear anchorage protein 1 (Anchorage... 37 0.095 sp|Q6FS63|MAD1_CANGA Spindle assembly checkpoint component ... 35 0.36 sp|O67358|TIG_AQUAE Trigger factor (TF) 35 0.47 sp|P39723|YAE7_YEAST Hypothetical 72.1 kDa protein in ACS1-... 34 0.80 sp|Q93Y94|RPO1_NICSY DNA-directed RNA polymerase 1, mitocho... 34 1.0 sp|Q27713|DRTS_PLABA Bifunctional dihydrofolate reductase-t... 34 1.0
>sp|Q9VCI3|LSD1_DROME Lipid storage droplets surface binding protein 1 Length = 431 Score = 55.1 bits (131), Expect = 4e-07 Identities = 64/269 (23%), Positives = 109/269 (40%), Gaps = 35/269 (13%) Frame = +3 Query: 33 ASSHSDNHFKSLDRVHHYPIVNDSLEYVLSYYQWIKSSNGILDNTLTRAEKTLKAVKDTA 212 A+S S H +++DR+ P+V S++ V + Y +K++N + AE T+ A +T Sbjct: 4 ATSGSGLHLEAIDRIGSIPLVESSVKRVETIYDKVKNNNRLFSWYFETAEATISAAYETI 63 Query: 213 EPIAVR----QLGFIDNAVLNQLDRFEKKCPIILEXXXXXXXXXXXXYHDSVESTKKTLM 380 +P AV+ + +DN + LD E++ P++ Y ++ E L+ Sbjct: 64 QP-AVKLFEPSIQRLDNVMCKSLDILEQRIPLV-------YLPPEMMYWNTKEYMSDHLV 115 Query: 381 WPVTKTLDTV---------SKTSDFAXXXXXXXXXXXMKIVLPATYLTNVSTNYVLDKTD 533 PV K D+V S + +A K V YL + T+ D+TD Sbjct: 116 RPVLKRADSVKQIGNAVLESPLTTYAAERIDGAFTVGDKFV--DKYLVPIQTDQ--DQTD 171 Query: 534 ------ELLDKYLP----------------DNNQCNEEPYSNADIVTKVKRLSGKTQQRL 647 L D + P D+N+ + + +R S K ++RL Sbjct: 172 VKKLVSALNDNFSPIRQQLNRMFGHKSPQEDDNEAVPDERGAIKAIHHGQRFSRKLKRRL 231 Query: 648 YKRASEQINALLKASLEKLHQIHVLIEEI 734 +R + AL K S E +H + E I Sbjct: 232 TQRTIAEARALKKQSKEAIHVLFYAAELI 260
>sp|Q9VXY7|LSD2_DROME Lipid storage droplets surface binding protein 2 (Lsd2) Length = 352 Score = 48.9 bits (115), Expect = 3e-05 Identities = 59/263 (22%), Positives = 95/263 (36%), Gaps = 44/263 (16%) Frame = +3 Query: 54 HFKSLDRVHHYPIVNDSLEYVLSYYQWIKSSNGILDNTLTRAEKTLKAVKDTAEPIAV-- 227 H +SL+R+ P+VN + + Y +K N + + LT AE + TA P Sbjct: 36 HLESLERIIKLPVVNAAWDKSQDVYGKVKGKNRVFEWALTAAEDCVTRAVTTAAPFVTKL 95 Query: 228 -RQLGFIDNAVLNQLDRFEKKCPIILEXXXXXXXXXXXXYHDSVESTKKTLMWPVTKTLD 404 R + ++D ++ +D+ E K PII + D V+ + ++ Sbjct: 96 DRPIAYVDQTLVKGIDKLEVKAPIIKDTPQEIYNQAKSKVIDVVQPHLERVVKFKAAGQQ 155 Query: 405 TVSKTSDFAXXXXXXXXXXXMKIVLPATY----LTNVSTNYVLDKTDELLDKYLP----- 557 + D A VL Y + V T L + LL+ Y P Sbjct: 156 KAASLKDLAWQKANE--------VLATQYGSLAVNGVDTTTAL--AERLLEYYFPKCESD 205 Query: 558 ---DNNQ-------------------CNEEPYSNADIVTKVKRLSGKTQQRLYKRASEQI 671 DN+ +E+P + V V RLS K +R+Y+ S QI Sbjct: 206 VEEDNDDKQNAVVQNGKSSENDMPVPASEDPVLHT--VQTVGRLSNKISRRVYRNVSRQI 263 Query: 672 NALLKASLE----------KLHQ 710 + K ++ KLHQ Sbjct: 264 KQVQKGNINDYLSSLIAALKLHQ 286
>sp|P43883|ADFP_MOUSE Adipophilin (Adipose differentiation-related protein) (ADRP) Length = 425 Score = 47.0 bits (110), Expect = 1e-04 Identities = 52/254 (20%), Positives = 94/254 (37%), Gaps = 23/254 (9%) Frame = +3 Query: 72 RVHHYPIVNDSLEYVLSYYQWIKSSNGILDNTLTRAEKTLKAVKDTAEPIAV-------R 230 RV + P+V+ + + V S Y K L + AEK +K V A A+ Sbjct: 15 RVANLPLVSSTYDLVSSAYVSTKDQYPYLRSVCEMAEKGVKTVTSAAMTSALPIIQKLEP 74 Query: 231 QLGFIDNAVLNQLDRFEKKCPIILEXXXXXXXXXXXXYHDSVESTKKTLMWPVTKTLDTV 410 Q+ + LDR E++ PI+ + + + T+ TV Sbjct: 75 QIAVANTYACKGLDRMEERLPILNQPTSEIVASARGAVTGAKDVVTTTMAGAKDSVASTV 134 Query: 411 SKTSDFAXXXXXXXXXXXMKI-------VLPATYLTNVSTNYVLDKTDELLDKYLP---D 560 S D + VL N + + K++ L+D+Y P + Sbjct: 135 SGVVDKTKGAVTGSVERTKSVVNGSINTVLGMVQFMNSGVDNAITKSEMLVDQYFPLTQE 194 Query: 561 NNQCNEEPYSNADIVTK------VKRLSGKTQQRLYKRASEQINALLKASLEKLHQIHVL 722 + + D+V K ++ LS K R Y +A ++ + S E + Q+H Sbjct: 195 ELEMEAKKVEGFDMVQKPSNYERLESLSTKLCSRAYHQALSRVKEAKQKSQETISQLHST 254 Query: 723 IEEINKRREDLHNS 764 + I R+++H++ Sbjct: 255 VHLIEFARKNMHSA 268
>sp|Q9TUM6|ADFP_BOVIN Adipophilin (Adipose differentiation-related protein) (ADRP) Length = 450 Score = 43.9 bits (102), Expect = 0.001 Identities = 57/270 (21%), Positives = 106/270 (39%), Gaps = 25/270 (9%) Frame = +3 Query: 30 VASSHSDNHFKSLDRVHHYPIVNDSLEYVLSYYQWIKSSNGILDNTLTRAEKTLKAVKDT 209 +AS + + RV + P+V+ + + V S Y K L + AEK +K + Sbjct: 1 MASVAVEPQLSVVTRVANLPLVSSTYDLVSSAYISRKDQYPYLKSLCEMAEKGMKTITSV 60 Query: 210 AEPIAVR-------QLGFIDNAVLNQLDRFEKKCPIILEXXXXXXXXXXXXY---HDSVE 359 A A+ Q+ + LDR E+K PI+ + D+V Sbjct: 61 AVTSALPIIQKLEPQIAVANTYACKGLDRIEEKLPILNQPTNQVVANAKGAMTGAKDAVT 120 Query: 360 ST----KKTLMWPVTKTLDTVSKTSDFAXXXXXXXXXXXMKIVLPATYLTNVSTNY--VL 521 +T K ++ +T +D + + VL + + +S+ L Sbjct: 121 TTVTGAKDSVASTITGVVDRTKGAVTGSVEKTKSVVSGSINTVLRSRVMQLMSSGVENAL 180 Query: 522 DKTDELLDKYLP---DNNQCNEEPYSNADIVTK------VKRLSGKTQQRLYKRASEQIN 674 K++ L+D+YLP D + + D+V K + LS K + R Y++A ++ Sbjct: 181 TKSELLVDQYLPLTKDELEKEAKKVEGFDMVQKPSYYVRLGSLSTKLRSRAYQQALCRVE 240 Query: 675 ALLKASLEKLHQIHVLIEEINKRREDLHNS 764 + E + Q+H R+++HN+ Sbjct: 241 EAKRKGQETISQLHSAFNLSELARKNVHNA 270
>sp|Q9N4M4|ANC1_CAEEL Nuclear anchorage protein 1 (Anchorage 1 protein) (Nesprin homolog) Length = 8545 Score = 37.4 bits (85), Expect = 0.095 Identities = 25/99 (25%), Positives = 51/99 (51%), Gaps = 1/99 (1%) Frame = +3 Query: 639 QRLYKRASEQINALLKASLEKLHQIHVLIEEINKRREDLHNSVAQEYSNTLDVIKKNNPF 818 +R Y+ +++NA + A +E L + +L E+++ +ED++N S +D + + Sbjct: 8289 ERQYEDTMDKLNAEITAEVELLRTLDILSNELSQCKEDINNP-----SVDVDELSRATML 8343 Query: 819 N-LINHSQIVSEALRYSKSLKNVVSSSTSVTLEYMVNNA 932 N I H + + S+ + V SSTS+ L+ ++ A Sbjct: 8344 NDAIAHLENQKVVVARSEKDRKFVESSTSIDLDQLLAEA 8382
>sp|Q6FS63|MAD1_CANGA Spindle assembly checkpoint component MAD1 (Mitotic arrest deficient protein 1) Length = 657 Score = 35.4 bits (80), Expect = 0.36 Identities = 44/202 (21%), Positives = 78/202 (38%) Frame = +3 Query: 504 STNYVLDKTDELLDKYLPDNNQCNEEPYSNADIVTKVKRLSGKTQQRLYKRASEQINALL 683 STN + D LLD Y + E D + + R G ++ R + IN Sbjct: 382 STNRNDAEYDSLLDNYKQETENLTNELKRLNDSLIEQNRNQGTEMKKRKLRNGDHININY 441 Query: 684 KASLEKLHQIHVLIEEINKRREDLHNSVAQEYSNTLDVIKKNNPFNLINHSQIVSEALRY 863 L +L Q ++ +E+ N++ V ++ LD ++K P N+ Q S L+ Sbjct: 442 SQRLNELQQNNLSLEKDNEKL----RLVVEKLEGKLDDLRKTKPKNIRILQQRDSPLLK- 496 Query: 864 SKSLKNVVSSSTSVTLEYMVNNALNGLDIVNDNLFKLQNFLQQARPESVTGELLIHTSTA 1043 L+++ L N NL + + + R ES L ++ Sbjct: 497 ---------------LQFVRRKENELLREENQNLLTMIENITENRNESFDKIPLTSYNSL 541 Query: 1044 VTEIRIALDSFIYHMKRYSRKK 1109 + I++D+ K++SR K Sbjct: 542 KYQYEISMDTNKKQEKKFSRLK 563
>sp|O67358|TIG_AQUAE Trigger factor (TF) Length = 478 Score = 35.0 bits (79), Expect = 0.47 Identities = 33/130 (25%), Positives = 61/130 (46%), Gaps = 5/130 (3%) Frame = +3 Query: 516 VLDKTDELLDKYLPDNNQCNEEPYSNADIVTKVKRLSGKTQQRLYKRASEQINALLKASL 695 V DK E+ D +P E + + +++ L T+ K+ E++ + +A++ Sbjct: 321 VADKLVEIHDIPVPQTLLRRELSFLVDRRLRELQALGIDTRYVDIKKIVEEVQPIAEANI 380 Query: 696 EKLHQIHVLIEE--INKRREDL---HNSVAQEYSNTLDVIKKNNPFNLINHSQIVSEALR 860 + + +E I ED+ + +A++Y T+D IKK F N Q+V E R Sbjct: 381 KLRFILDKYAQEKGIEPTGEDIEAQYKELAEQYGTTVDEIKKY--FKENNLEQVVYEDAR 438 Query: 861 YSKSLKNVVS 890 K+LK ++S Sbjct: 439 RKKALKEIIS 448
>sp|P39723|YAE7_YEAST Hypothetical 72.1 kDa protein in ACS1-GCV3 intergenic region Length = 622 Score = 34.3 bits (77), Expect = 0.80 Identities = 34/142 (23%), Positives = 56/142 (39%), Gaps = 12/142 (8%) Frame = +3 Query: 606 TKVKRLSGKTQQRLYKRASEQINALLKASLEKLHQIHVLIEEINKRREDLHNSVAQEYSN 785 TK+ +L +QIN L ++K + HV+ +++K DL N + N Sbjct: 401 TKINKLKENNWDSYINDLEKQINDL---QIDKSEEFHVIQNQLDKL--DLENYQLKNQLN 455 Query: 786 TLD------------VIKKNNPFNLINHSQIVSEALRYSKSLKNVVSSSTSVTLEYMVNN 929 TLD IK N NL+ H + L+ ++ + V N Sbjct: 456 TLDNQKLILSQYESNFIKFNQ--NLLLHLDSIFNILQKILQESSIAQFDRKMKSIKSVPN 513 Query: 930 ALNGLDIVNDNLFKLQNFLQQA 995 AL L+++ L L F++ A Sbjct: 514 ALKNLNLIQPKLESLYTFIETA 535
>sp|Q93Y94|RPO1_NICSY DNA-directed RNA polymerase 1, mitochondrial precursor (T7 bacteriophage-type single subunit RNA polymerase 1) (NsRpoT-A) Length = 1002 Score = 33.9 bits (76), Expect = 1.0 Identities = 31/126 (24%), Positives = 53/126 (42%), Gaps = 7/126 (5%) Frame = +3 Query: 642 RLYKRASEQINALLKASLEKLHQIHVLIEEINKRREDLHNSVAQEYSNTLDVIKKNNPFN 821 R Y A+E A+ S E + +I LIEE+NK E L ++ + T+ + +N Sbjct: 112 RSYGSAAE---AIASTSEEDIDEIQELIEEMNKENEALKTNLQPKQPKTIGGMGVGK-YN 167 Query: 822 LINHSQIVSE-------ALRYSKSLKNVVSSSTSVTLEYMVNNALNGLDIVNDNLFKLQN 980 L+ QI E A Y + L ++ + L YM + L + + D + Q Sbjct: 168 LLRRRQIKVETEAWEEAAKEYQELLMDMCEQKLAPNLPYMKSLFLGWFEPLRDAIAAEQK 227 Query: 981 FLQQAR 998 + + Sbjct: 228 LCDEGK 233
>sp|Q27713|DRTS_PLABA Bifunctional dihydrofolate reductase-thymidylate synthase (DHFR-TS) [Includes: Dihydrofolate reductase ; Thymidylate synthase ] Length = 587 Score = 33.9 bits (76), Expect = 1.0 Identities = 40/179 (22%), Positives = 78/179 (43%), Gaps = 27/179 (15%) Frame = +3 Query: 585 YSNADIVTKVKRLSGKTQQRLYKRASEQINALLKASLEKLHQIHVL----IEEINKRRED 752 Y N + ++K K ++ + + ++N + +S L I V+ E I K+ + Sbjct: 61 YINENNYIRLKWKRDKYMEKHNLKNNVELNTNIISSTNNLQNIVVMGKKSWESIPKKFKP 120 Query: 753 LHNSVAQEYSNTL---DVIKKNNPFN----LINHSQIVSEALRYSKSLKNVVSSSTSVTL 911 L N + S TL D++ +NN N +I + L+ +K K + +SV Sbjct: 121 LQNRINIILSRTLKKEDIVNENNNENNNVIIIKSVDDLFPILKCTKYYKCFIIGGSSVYK 180 Query: 912 EYM------------VNNALNGLDI----VNDNLFKLQNFLQQARPESVTGELLIHTST 1040 E++ +NN+ N D+ +N+NLFK+ + + T + +I++ T Sbjct: 181 EFLDRNLIKKIYFTRINNSYN-CDVLFPEINENLFKITSISDVYYSNNTTLDFIIYSKT 238
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 128,622,138
Number of Sequences: 369166
Number of extensions: 2461845
Number of successful extensions: 7976
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7506
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7963
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 14908696450
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail