DrC_00556
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
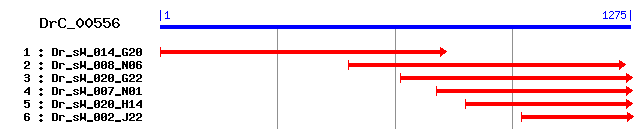
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00556 (1272 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9H6R6|ZDHC6_HUMAN Probable palmitoyltransferase ZDHHC6 ... 301 2e-81 sp|Q9CPV7|ZDHC6_MOUSE Probable palmitoyltransferase ZDHHC6 ... 296 7e-80 sp|Q4PE27|PFA4_USTMA Palmitoyltransferase PFA4 (Protein fat... 122 3e-27 sp|Q9ESG8|ZDH16_MOUSE Probable palmitoyltransferase ZDHHC16... 108 4e-23 sp|Q5KLN1|PFA4_CRYNE Palmitoyltransferase PFA4 (Protein fat... 108 4e-23 sp|Q58CU4|ZDH16_BOVIN Probable palmitoyltransferase ZDHHC16... 103 8e-22 sp|Q969W1|ZDH16_HUMAN Probable palmitoyltransferase ZDHHC16... 102 3e-21 sp|Q4R7E2|ZDH16_MACFA Probable palmitoyltransferase ZDHHC16... 102 3e-21 sp|Q5BD15|PFA4_EMENI Palmitoyltransferase PFA4 (Protein fat... 97 1e-19 sp|Q6BLY8|PFA4_DEBHA Palmitoyltransferase PFA4 (Protein fat... 93 1e-18
>sp|Q9H6R6|ZDHC6_HUMAN Probable palmitoyltransferase ZDHHC6 (Zinc finger DHHC domain-containing protein 6) (DHHC-6) (Zinc finger protein 376) (Transmembrane protein H4) sp|Q5REH2|ZDHC6_PONPY Probable palmitoyltransferase ZDHHC6 (Zinc finger DHHC domain-containing protein 6) (DHHC-6) Length = 413 Score = 301 bits (772), Expect = 2e-81 Identities = 148/362 (40%), Positives = 213/362 (58%), Gaps = 13/362 (3%) Frame = +3 Query: 12 IHFSIYVYEFYLVVNNYLNASFIGPGFVNLGWEPKLEDDKQFLQYCELCEGYKPPRSHHC 191 ++F + + +++ NY NA F+GPGFV LGW+P++ D +LQYC++C+ YK PRSHHC Sbjct: 56 VNFIMLINWTVMILYNYFNAMFVGPGFVPLGWKPEISQDTMYLQYCKVCQAYKAPRSHHC 115 Query: 192 RKCDRCVMKMDHHCPWINTCCGHLNHGYFVYFLFFAPIGCIHSTYILGLTLYYAIYVDYF 371 RKC+RCVMKMDHHCPWIN CCG+ NH F FL AP+GCIH+ +I +T+Y +Y Sbjct: 116 RKCNRCVMKMDHHCPWINNCCGYQNHASFTLFLLLAPLGCIHAAFIFVMTMYTQLYHRLS 175 Query: 372 FNF------------DDQYIVILSLMEFLCSFFXXXXXXXXXXXXXSLWVIQMRSIMKNE 515 F + D IV L F + F L+ IQM+ I++N+ Sbjct: 176 FGWNTVKIDMSAARRDPLPIVPFGLAAFATTLFALGLALGTTIAVGMLFFIQMKIILRNK 235 Query: 516 TGVESWIKEKSQLRIQENNNSNERFVYPYDLG-RIRNMKEILTWSGRCRGDGITWTVKDG 692 T +ESWI+EK++ RIQ +E FV+PYD+G R RN K++ TWSG GDG+ W V++G Sbjct: 236 TSIESWIEEKAKDRIQ-YYQLDEVFVFPYDMGSRWRNFKQVFTWSGVPEGDGLEWPVREG 294 Query: 693 CGQYDLTIEQIKQKRDKVLHSQNYEIRRSYNGGLFPVISFGCRVVYGSPCTDENRLPVKP 872 C QY LTIEQ+KQK DK + S Y++ Y+G P ++ G + + SPCT+E R+ ++ Sbjct: 295 CHQYSLTIEQLKQKADKRVRSVRYKVIEDYSGACCP-LNKGIKTFFTSPCTEEPRIQLQK 353 Query: 873 GQIVMVTRWRKHWLYGQILXXXXXXXXXXXXXXXXVESSQQRRVATKRANSRGWFPAVCA 1052 G+ ++ TR ++WLYG + ++ S V + RGWFP C Sbjct: 354 GEFILATRGLRYWLYGDKI----------------LDDSFIEGV----SRIRGWFPRKCV 393 Query: 1053 RQ 1058 + Sbjct: 394 EK 395
>sp|Q9CPV7|ZDHC6_MOUSE Probable palmitoyltransferase ZDHHC6 (Zinc finger DHHC domain-containing protein 6) (DHHC-6) (H4 homolog) Length = 413 Score = 296 bits (759), Expect = 7e-80 Identities = 148/385 (38%), Positives = 214/385 (55%), Gaps = 13/385 (3%) Frame = +3 Query: 12 IHFSIYVYEFYLVVNNYLNASFIGPGFVNLGWEPKLEDDKQFLQYCELCEGYKPPRSHHC 191 ++F + + +++ NY NA F GPGFV GW+P+ D +LQYC++C+ YK PRSHHC Sbjct: 56 VNFIMLINWTVMILYNYFNAMFAGPGFVPRGWKPEKSQDSMYLQYCKVCQAYKAPRSHHC 115 Query: 192 RKCDRCVMKMDHHCPWINTCCGHLNHGYFVYFLFFAPIGCIHSTYILGLTLYYAIYVDYF 371 RKC+RCVMKMDHHCPWIN CCGH NH F FL AP+GC H+ +I +T+Y +Y Sbjct: 116 RKCNRCVMKMDHHCPWINNCCGHQNHASFTLFLLLAPLGCTHAAFIFVMTMYTQLYNRLS 175 Query: 372 FNF------------DDQYIVILSLMEFLCSFFXXXXXXXXXXXXXSLWVIQMRSIMKNE 515 F + D IV L F + F L+ IQ++ I++N+ Sbjct: 176 FGWNTVKIDMSAARRDPPPIVPFGLAAFAATLFALGLALGTTIAVGMLFFIQIKIILRNK 235 Query: 516 TGVESWIKEKSQLRIQENNNSNERFVYPYDLG-RIRNMKEILTWSGRCRGDGITWTVKDG 692 T +ESWI+EK++ RIQ +E F++PYD+G + +N K++ TWSG GDG+ W +++G Sbjct: 236 TSIESWIEEKAKDRIQ-YYQLDEVFIFPYDMGSKWKNFKQVFTWSGVPEGDGLEWPIREG 294 Query: 693 CGQYDLTIEQIKQKRDKVLHSQNYEIRRSYNGGLFPVISFGCRVVYGSPCTDENRLPVKP 872 C QY LTIEQ+KQK DK + S Y++ YNG P ++ G R + SPCT+E R+ ++ Sbjct: 295 CDQYSLTIEQLKQKADKRVRSVRYKVIEDYNGACCP-LNRGVRTFFTSPCTEEPRIRLQK 353 Query: 873 GQIVMVTRWRKHWLYGQILXXXXXXXXXXXXXXXXVESSQQRRVATKRANSRGWFPAVCA 1052 G+ ++ TR ++WLYG + +E + + RGWFP C Sbjct: 354 GEFILATRGLRYWLYGDKI-----------LDDSFIEGT---------SRVRGWFPRNCV 393 Query: 1053 RQKYLGDEGGTAVPQKADRNGDSQP 1127 +K +GDS P Sbjct: 394 --------------EKCPCDGDSDP 404
>sp|Q4PE27|PFA4_USTMA Palmitoyltransferase PFA4 (Protein fatty acyltransferase 4) Length = 604 Score = 122 bits (305), Expect = 3e-27 Identities = 65/210 (30%), Positives = 106/210 (50%), Gaps = 12/210 (5%) Frame = +3 Query: 108 EPKLEDDKQFLQ--YCELCEGYKPPRSHHCRKCDRCVMKMDHHCPWINTCCGHLNHGYFV 281 EP LE + + YC+ C +KPPRSHHC+ C RCV++MDHHCPW+ C GH NH +F+ Sbjct: 168 EPSLELKQAIYRPRYCKTCSAFKPPRSHHCKTCQRCVLRMDHHCPWLANCVGHFNHAHFI 227 Query: 282 YFLFFAPIGCIHSTYILGLTLYYAIYVDYFFNFDDQYIVILSLMEFLCSFFXXXXXXXXX 461 FLF+ + C++ ++ + +D F ++ + +L + Sbjct: 228 RFLFYVDVTCLYHLIMISCRV-----LDSFNSYTYWREPCARELVWLVVNYALCIPVILL 282 Query: 462 XXXXSLWVIQMRSIMKNETGVESWIKEKSQLRIQENNNSNERFVYPYDLGRIRNMKEI-- 635 SL+ ++ N+T +ESW K+++ I+ + YPYDLG RN++++ Sbjct: 283 VGIFSLYHFYCLAV--NQTTIESWEKDRTATMIRRGR--VRKVKYPYDLGLWRNVRQVLG 338 Query: 636 ---LTW-----SGRCRGDGITWTVKDGCGQ 701 L W R GDG+ + V +G G+ Sbjct: 339 ASPLVWCLPGAGARMAGDGLKYPVANGLGK 368
>sp|Q9ESG8|ZDH16_MOUSE Probable palmitoyltransferase ZDHHC16 (Zinc finger DHHC domain-containing protein 16) (DHHC-16) (Abl-philin 2) Length = 361 Score = 108 bits (269), Expect = 4e-23 Identities = 71/217 (32%), Positives = 102/217 (47%), Gaps = 9/217 (4%) Frame = +3 Query: 15 HFSIYVYEFYLVVNNYLNASFIGPGFVNLGWEPKLEDDKQFLQYCELCEGYKPPRSHHCR 194 HF + L+V +Y A PG+ P+ +D + C+ C KP R+HHC Sbjct: 118 HFFYSHWNLILIVFHYYQAITTPPGY-----PPQGRNDIATVSICKKCIYPKPARTHHCS 172 Query: 195 KCDRCVMKMDHHCPWINTCCGHLNHGYFVYFLFFAPIGCIHSTY---ILGLTLYYAIYVD 365 C+RCV+KMDHHCPW+N C GH NH YF F FF +GC++ +Y L Y AI Sbjct: 173 ICNRCVLKMDHHCPWLNNCVGHYNHRYFFSFCFFMTLGCVYCSYGSWDLFREAYAAIETY 232 Query: 366 Y-----FFNFDDQYI-VILSLMEFLCSFFXXXXXXXXXXXXXSLWVIQMRSIMKNETGVE 527 + F+F ++ L + FLCS ++W + I + ET +E Sbjct: 233 HQTPPPTFSFRERITHKSLVYLWFLCS------SVALALGALTMWHAVL--ISRGETSIE 284 Query: 528 SWIKEKSQLRIQENNNSNERFVYPYDLGRIRNMKEIL 638 I +K + R+Q F PY+ G + N K L Sbjct: 285 RHINKKERRRLQAKGRV---FRNPYNYGCLDNWKVFL 318
>sp|Q5KLN1|PFA4_CRYNE Palmitoyltransferase PFA4 (Protein fatty acyltransferase 4) Length = 456 Score = 108 bits (269), Expect = 4e-23 Identities = 65/236 (27%), Positives = 109/236 (46%), Gaps = 15/236 (6%) Frame = +3 Query: 42 YLVVNNYLNASFIGPGFVNLGWEPKL--------EDDKQFLQYCELCEGYKPPRSHHCRK 197 +++ NY PG V GW P + + +YC+ CE YKPPR+HHCR+ Sbjct: 54 FMIFWNYRLCVITSPGSVPEGWRPNIGAMDGMEVKKGTHTPRYCKNCEHYKPPRAHHCRQ 113 Query: 198 CDRCVMKMDHHCPWINTCCGHLNHGYFVYFLFFAPIGCIHSTYILGLTLYYAIYVDYFFN 377 C C + +HCPWI C G N G+F+ FL + IG +T+ L + + +Y+ +++ Sbjct: 114 CKTCWV---NHCPWIGNCVGFYNQGHFIRFLLWVDIG---TTFHLIIMVRRVLYIAEYYH 167 Query: 378 FDDQYIVILSLMEFLCSFFXXXXXXXXXXXXXSLWVIQMRSIMKNETGVESWIKEKSQLR 557 + +L FL F S++ + + N T +E W K+K Sbjct: 168 QEPTLADVL----FLVFNFATCVPVWLCVGMFSIYHVYL--ACGNSTTIEGWEKDKVATL 221 Query: 558 IQENNNSNERFVYPYDLGRIRNMKEIL-------TWSGRCRGDGITWTVKDGCGQY 704 I+ + YPY++G +N+K +L W + +GDG+++ V G + Sbjct: 222 IRRGKIKEVK--YPYNIGIYKNIKSVLGPNPFLWLWPQKMQGDGLSFPVNPSAGDH 275
>sp|Q58CU4|ZDH16_BOVIN Probable palmitoyltransferase ZDHHC16 (Zinc finger DHHC domain-containing protein 16) (DHHC-16) Length = 377 Score = 103 bits (258), Expect = 8e-22 Identities = 44/104 (42%), Positives = 60/104 (57%) Frame = +3 Query: 15 HFSIYVYEFYLVVNNYLNASFIGPGFVNLGWEPKLEDDKQFLQYCELCEGYKPPRSHHCR 194 HF + L+V +Y A PG+ P+ +D + C+ C KP R+HHC Sbjct: 118 HFFYSHWNLILIVFHYYQAITTPPGY-----PPQGRNDMTTVSICKKCINPKPARTHHCS 172 Query: 195 KCDRCVMKMDHHCPWINTCCGHLNHGYFVYFLFFAPIGCIHSTY 326 C+RCV+KMDHHCPW+N C GH NH YF F FF +GC++ +Y Sbjct: 173 ICNRCVLKMDHHCPWLNNCVGHYNHRYFFSFCFFMTLGCVYCSY 216
>sp|Q969W1|ZDH16_HUMAN Probable palmitoyltransferase ZDHHC16 (Zinc finger DHHC domain-containing protein 16) (DHHC-16) Length = 377 Score = 102 bits (253), Expect = 3e-21 Identities = 44/104 (42%), Positives = 60/104 (57%) Frame = +3 Query: 15 HFSIYVYEFYLVVNNYLNASFIGPGFVNLGWEPKLEDDKQFLQYCELCEGYKPPRSHHCR 194 HF + L+V +Y A PG+ P+ +D + C+ C KP R+HHC Sbjct: 118 HFFYSHWNLILIVFHYYQAITTPPGY-----PPQGRNDIATVSICKKCIYPKPARTHHCS 172 Query: 195 KCDRCVMKMDHHCPWINTCCGHLNHGYFVYFLFFAPIGCIHSTY 326 C+RCV+KMDHHCPW+N C GH NH YF F FF +GC++ +Y Sbjct: 173 ICNRCVLKMDHHCPWLNNCVGHYNHRYFFSFCFFMTLGCVYCSY 216
>sp|Q4R7E2|ZDH16_MACFA Probable palmitoyltransferase ZDHHC16 (Zinc finger DHHC domain-containing protein 16) (DHHC-16) Length = 377 Score = 102 bits (253), Expect = 3e-21 Identities = 44/104 (42%), Positives = 60/104 (57%) Frame = +3 Query: 15 HFSIYVYEFYLVVNNYLNASFIGPGFVNLGWEPKLEDDKQFLQYCELCEGYKPPRSHHCR 194 HF + L+V +Y A PG+ P+ +D + C+ C KP R+HHC Sbjct: 118 HFFYSHWNLILIVFHYYQAITTPPGY-----PPQGRNDIATVSICKKCIYPKPARTHHCS 172 Query: 195 KCDRCVMKMDHHCPWINTCCGHLNHGYFVYFLFFAPIGCIHSTY 326 C+RCV+KMDHHCPW+N C GH NH YF F FF +GC++ +Y Sbjct: 173 ICNRCVLKMDHHCPWLNNCVGHYNHRYFFSFCFFMTLGCVYCSY 216
>sp|Q5BD15|PFA4_EMENI Palmitoyltransferase PFA4 (Protein fatty acyltransferase 4) Length = 435 Score = 97.1 bits (240), Expect = 1e-19 Identities = 41/89 (46%), Positives = 54/89 (60%), Gaps = 6/89 (6%) Frame = +3 Query: 60 YLNASFIGPGFVNLGWEP----KLEDDKQF--LQYCELCEGYKPPRSHHCRKCDRCVMKM 221 Y A + PG V GW P +L+ D+ ++C CE YKPPR+HHC+ C+RCV KM Sbjct: 58 YYRACTVDPGHVPKGWMPSDRERLKADRASGRQRWCRRCEAYKPPRAHHCKTCERCVPKM 117 Query: 222 DHHCPWINTCCGHLNHGYFVYFLFFAPIG 308 DHHCPW + C H +F FLF+A +G Sbjct: 118 DHHCPWTSNCVSHFTFPHFARFLFYAVVG 146
>sp|Q6BLY8|PFA4_DEBHA Palmitoyltransferase PFA4 (Protein fatty acyltransferase 4) Length = 402 Score = 93.2 bits (230), Expect = 1e-18 Identities = 41/104 (39%), Positives = 63/104 (60%), Gaps = 2/104 (1%) Frame = +3 Query: 30 VYEFYLVVN--NYLNASFIGPGFVNLGWEPKLEDDKQFLQYCELCEGYKPPRSHHCRKCD 203 +YE Y+ + +Y A + PG + PK ++ ++C+ C+ YKP RSHHC+ C+ Sbjct: 42 LYEVYVCIVWLSYYLAIVVDPGSPPKNFTPKA---GEWRRWCKKCQNYKPERSHHCKTCN 98 Query: 204 RCVMKMDHHCPWINTCCGHLNHGYFVYFLFFAPIGCIHSTYILG 335 +CV+KMDHHCPW C GH N +F+ F+FF +G + + LG Sbjct: 99 KCVLKMDHHCPWTYNCVGHNNLPHFLRFVFFLIVGMTYVLFQLG 142
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 153,333,218
Number of Sequences: 369166
Number of extensions: 3372532
Number of successful extensions: 10793
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9744
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10659
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 14718776750
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail