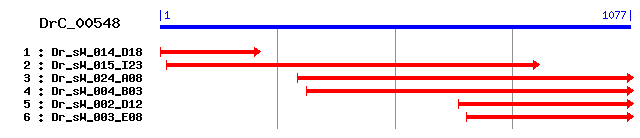
DrC_00548
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00548 (1077 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q8BFR5|EFTU_MOUSE Elongation factor Tu, mitochondrial pr... 385 e-107 sp|P49410|EFTU_BOVIN Elongation factor Tu, mitochondrial pr... 381 e-105 sp|P49411|EFTU_HUMAN Elongation factor Tu, mitochondrial pr... 380 e-105 sp|P48865|EFTU_RICPR Elongation factor Tu (EF-Tu) 357 2e-98 sp|Q8KT95|EFTU_RICTY Elongation factor Tu (EF-Tu) 356 7e-98 sp|P42473|EFTU_CHLVI Elongation factor Tu (EF-Tu) 355 9e-98 sp|P42480|EFTU_TAXOC Elongation factor Tu (EF-Tu) 355 1e-97 sp|Q9Y700|EFTU_SCHPO Elongation factor Tu, mitochondrial pr... 355 1e-97 sp|O50306|EFTU_BACST Elongation factor Tu (EF-Tu) 355 1e-97 sp|Q8KT99|EFTU_RICHE Elongation factor Tu (EF-Tu) 355 2e-97
>sp|Q8BFR5|EFTU_MOUSE Elongation factor Tu, mitochondrial precursor Length = 452 Score = 385 bits (990), Expect = e-107 Identities = 189/336 (56%), Positives = 255/336 (75%), Gaps = 3/336 (0%) Frame = +1 Query: 1 KRRGITINAAVVDYSTDARHYAHTDCPGHADYIKNMITGANQMEAAILVVAATDGSMPQT 180 + RGITINAA V+YST ARHYAHTDCPGHADY+KNMITG ++ ILVVAA DG MPQT Sbjct: 102 RARGITINAAHVEYSTAARHYAHTDCPGHADYVKNMITGTAPLDGCILVVAANDGPMPQT 161 Query: 181 REHLVLAKQIGIDKLVVFINKADAA-DNEMIELVEMEVRDLLTKYGYDGENIPFVTGSAL 357 REHL+LAKQIG++ +VV++NKADA D+EM+ELVE+E+R+LLT++GY GE P + GSAL Sbjct: 162 REHLLLAKQIGVEHVVVYVNKADAVQDSEMVELVELEIRELLTEFGYKGEETPVIVGSAL 221 Query: 358 LALEGKNEEIGKNKIIELLNKIDE-IPLPAREIDKPLFLPVEHVLSITGRGTVCTGRVER 534 ALE ++ E+G + +LL+ +D IP+P R++DKP LPVE V SI GRGTV TG +ER Sbjct: 222 CALEQRDPELGVKSVQKLLDAVDTYIPVPTRDLDKPFLLPVESVYSIPGRGTVVTGTLER 281 Query: 535 GVINLGLAVEIIGYNASVKTTITGIEMFRQLLDKAVPGDQVGVLLRSVKKEELRRGMVII 714 G++ G E++G+N +++T +TGIEMF + L++A GD +G L+R +K+E+LRRG+V++ Sbjct: 282 GILKKGDECELLGHNKNIRTVVTGIEMFHKSLERAEAGDNLGALVRGLKREDLRRGLVMV 341 Query: 715 SPKSVQMYNYVEAQIYLLTEAEGGRKKPSTKNFQSQIYSKSWDCPAFTILPEGKELIMPG 894 P S+Q + VEAQ+Y+L++ EGGR KP +F ++S +WD ILP GKEL MPG Sbjct: 342 KPGSIQPHQKVEAQVYILSKEEGGRHKPFVSHFMPVMFSLTWDMACRVILPPGKELAMPG 401 Query: 895 EDSSVKLHLQKKMVLEVGQRFTLRCG-ITLGYGVVT 999 ED + L L++ M+LE GQRFTLR G T+G G+VT Sbjct: 402 EDLKLSLILRQPMILEKGQRFTLRDGNKTIGTGLVT 437
>sp|P49410|EFTU_BOVIN Elongation factor Tu, mitochondrial precursor (EF-Tu) Length = 452 Score = 381 bits (979), Expect = e-105 Identities = 188/336 (55%), Positives = 253/336 (75%), Gaps = 3/336 (0%) Frame = +1 Query: 1 KRRGITINAAVVDYSTDARHYAHTDCPGHADYIKNMITGANQMEAAILVVAATDGSMPQT 180 + RGITINAA V+YST ARHYAHTDCPGHADY+KNMITG ++ ILVVAA DG MPQT Sbjct: 102 RARGITINAAHVEYSTAARHYAHTDCPGHADYVKNMITGTAPLDGCILVVAANDGPMPQT 161 Query: 181 REHLVLAKQIGIDKLVVFINKADAA-DNEMIELVEMEVRDLLTKYGYDGENIPFVTGSAL 357 REHL+LA+QIG++ +VV++NKADA D+EM+ELVE+E+R+LLT++GY GE P + GSAL Sbjct: 162 REHLLLARQIGVEHVVVYVNKADAVQDSEMVELVELEIRELLTEFGYKGEETPIIVGSAL 221 Query: 358 LALEGKNEEIGKNKIIELLNKIDE-IPLPAREIDKPLFLPVEHVLSITGRGTVCTGRVER 534 ALE ++ E+G + +LL+ +D IP+P R+++KP LPVE V SI GRGTV TG +ER Sbjct: 222 CALEQRDPELGLKSVQKLLDAVDTYIPVPTRDLEKPFLLPVESVYSIPGRGTVVTGTLER 281 Query: 535 GVINLGLAVEIIGYNASVKTTITGIEMFRQLLDKAVPGDQVGVLLRSVKKEELRRGMVII 714 G++ G E +G++ +++T +TGIEMF + LD+A GD +G L+R +K+E+LRRG+V+ Sbjct: 282 GILKKGDECEFLGHSKNIRTVVTGIEMFHKSLDRAEAGDNLGALVRGLKREDLRRGLVMA 341 Query: 715 SPKSVQMYNYVEAQIYLLTEAEGGRKKPSTKNFQSQIYSKSWDCPAFTILPEGKELIMPG 894 P S+Q + VEAQ+Y+LT+ EGGR KP +F ++S +WD ILP GKEL MPG Sbjct: 342 KPGSIQPHQKVEAQVYILTKEEGGRHKPFVSHFMPVMFSLTWDMACRIILPPGKELAMPG 401 Query: 895 EDSSVKLHLQKKMVLEVGQRFTLRCG-ITLGYGVVT 999 ED + L L++ M+LE GQRFTLR G T+G G+VT Sbjct: 402 EDLKLTLILRQPMILEKGQRFTLRDGNRTIGTGLVT 437
>sp|P49411|EFTU_HUMAN Elongation factor Tu, mitochondrial precursor (EF-Tu) (P43) Length = 452 Score = 380 bits (976), Expect = e-105 Identities = 186/339 (54%), Positives = 256/339 (75%), Gaps = 3/339 (0%) Frame = +1 Query: 1 KRRGITINAAVVDYSTDARHYAHTDCPGHADYIKNMITGANQMEAAILVVAATDGSMPQT 180 + RGITINAA V+YST ARHYAHTDCPGHADY+KNMITG ++ ILVVAA DG MPQT Sbjct: 102 RARGITINAAHVEYSTAARHYAHTDCPGHADYVKNMITGTAPLDGCILVVAANDGPMPQT 161 Query: 181 REHLVLAKQIGIDKLVVFINKADAA-DNEMIELVEMEVRDLLTKYGYDGENIPFVTGSAL 357 REHL+LA+QIG++ +VV++NKADA D+EM+ELVE+E+R+LLT++GY GE P + GSAL Sbjct: 162 REHLLLARQIGVEHVVVYVNKADAVQDSEMVELVELEIRELLTEFGYKGEETPVIVGSAL 221 Query: 358 LALEGKNEEIGKNKIIELLNKIDE-IPLPAREIDKPLFLPVEHVLSITGRGTVCTGRVER 534 ALEG++ E+G + +LL+ +D IP+PAR+++KP LPVE V S+ GRGTV TG +ER Sbjct: 222 CALEGRDPELGLKSVQKLLDAVDTYIPVPARDLEKPFLLPVEAVYSVPGRGTVVTGTLER 281 Query: 535 GVINLGLAVEIIGYNASVKTTITGIEMFRQLLDKAVPGDQVGVLLRSVKKEELRRGMVII 714 G++ G E++G++ +++T +TGIEMF + L++A GD +G L+R +K+E+LRRG+V++ Sbjct: 282 GILKKGDECELLGHSKNIRTVVTGIEMFHKSLERAEAGDNLGALVRGLKREDLRRGLVMV 341 Query: 715 SPKSVQMYNYVEAQIYLLTEAEGGRKKPSTKNFQSQIYSKSWDCPAFTILPEGKELIMPG 894 P S++ + VEAQ+Y+L++ EGGR KP +F ++S +WD ILP KEL MPG Sbjct: 342 KPGSIKPHQKVEAQVYILSKEEGGRHKPFVSHFMPVMFSLTWDMACRIILPPEKELAMPG 401 Query: 895 EDSSVKLHLQKKMVLEVGQRFTLRCG-ITLGYGVVTKLL 1008 ED L L++ M+LE GQRFTLR G T+G G+VT L Sbjct: 402 EDLKFNLILRQPMILEKGQRFTLRDGNRTIGTGLVTNTL 440
>sp|P48865|EFTU_RICPR Elongation factor Tu (EF-Tu) Length = 394 Score = 357 bits (917), Expect = 2e-98 Identities = 177/338 (52%), Positives = 245/338 (72%), Gaps = 3/338 (0%) Frame = +1 Query: 1 KRRGITINAAVVDYSTDARHYAHTDCPGHADYIKNMITGANQMEAAILVVAATDGSMPQT 180 K RGITI+ A V+Y T RHYAH DCPGHADY+KNMITGA QM+ AILVV+A DG MPQT Sbjct: 57 KERGITISTAHVEYETQNRHYAHVDCPGHADYVKNMITGAAQMDGAILVVSAADGPMPQT 116 Query: 181 REHLVLAKQIGIDKLVVFINKADAADN-EMIELVEMEVRDLLTKYGYDGENIPFVTGSAL 357 REH++LAKQ+G+ +VVF+NK D D+ +++ELVEMEVR+LL+KYG+ G IP + GSAL Sbjct: 117 REHILLAKQVGVPAMVVFLNKVDMVDDPDLLELVEMEVRELLSKYGFPGNEIPIIKGSAL 176 Query: 358 LALEGKNEEIGKNKIIELLNKIDE-IPLPAREIDKPLFLPVEHVLSITGRGTVCTGRVER 534 ALEGK E G+ I EL+N +D IP P R DKP +P+E V SI+GRGTV TGRVE Sbjct: 177 QALEGKPE--GEKAINELMNAVDSYIPQPIRATDKPFLMPIEDVFSISGRGTVVTGRVES 234 Query: 535 GVINLGLAVEIIGYNASVKTTITGIEMFRQLLDKAVPGDQVGVLLRSVKKEELRRGMVII 714 G+I +G +EI+G + KTT TG+EMFR+LLD+ GD VG+LLR K+EE+ RG V+ Sbjct: 235 GIIKVGEEIEIVGLKNTQKTTCTGVEMFRKLLDEGQSGDNVGILLRGTKREEVERGQVLA 294 Query: 715 SPKSVQMYNYVEAQIYLLTEAEGGRKKPSTKNFQSQIYSKSWDCPAFTILPEGKELIMPG 894 P S++ ++ EA++Y+L++ EGGR P T +++ Q Y ++ D LP K+++MPG Sbjct: 295 KPGSIKPHDKFEAEVYVLSKEEGGRHTPFTNDYRPQFYFRTTDVTGTIKLPSDKQMVMPG 354 Query: 895 EDSSVKLHLQKKMVLEVGQRFTLR-CGITLGYGVVTKL 1005 ++++ + L K + ++ G +F++R G T+G G+VTK+ Sbjct: 355 DNATFSVELIKPIAMQEGLKFSIREGGRTVGAGIVTKI 392
>sp|Q8KT95|EFTU_RICTY Elongation factor Tu (EF-Tu) Length = 394 Score = 356 bits (913), Expect = 7e-98 Identities = 177/338 (52%), Positives = 246/338 (72%), Gaps = 3/338 (0%) Frame = +1 Query: 1 KRRGITINAAVVDYSTDARHYAHTDCPGHADYIKNMITGANQMEAAILVVAATDGSMPQT 180 K RGITI+ A V+Y T+ RHYAH DCPGHADY+KNMITGA QM+ AILVV+A DG MPQT Sbjct: 57 KERGITISTAHVEYETNNRHYAHVDCPGHADYVKNMITGAAQMDGAILVVSAADGPMPQT 116 Query: 181 REHLVLAKQIGIDKLVVFINKADAADN-EMIELVEMEVRDLLTKYGYDGENIPFVTGSAL 357 REH++LAKQ+G+ +VVF+NK D D+ +++ELVEMEVR+LL+KYG+ G IP + GSAL Sbjct: 117 REHILLAKQVGVPAMVVFLNKVDMVDDPDLLELVEMEVRELLSKYGFPGNEIPIIKGSAL 176 Query: 358 LALEGKNEEIGKNKIIELLNKIDE-IPLPAREIDKPLFLPVEHVLSITGRGTVCTGRVER 534 ALEGK E G+ I EL+N +D IP P R DKP +P+E V SI+GRGTV TGRVE Sbjct: 177 QALEGKPE--GEKAINELMNAVDSYIPQPIRATDKPFLMPIEDVFSISGRGTVVTGRVES 234 Query: 535 GVINLGLAVEIIGYNASVKTTITGIEMFRQLLDKAVPGDQVGVLLRSVKKEELRRGMVII 714 G+I +G +EI+G + KTT TG+EMFR+LLD+ GD VG+LLR K+EE+ RG V+ Sbjct: 235 GIIKVGEEIEIVGLKNTQKTTCTGVEMFRKLLDEGQSGDNVGILLRGTKREEVERGQVLA 294 Query: 715 SPKSVQMYNYVEAQIYLLTEAEGGRKKPSTKNFQSQIYSKSWDCPAFTILPEGKELIMPG 894 P S++ ++ EA++Y+L++ EGGR P T +++ Q Y ++ D LP K+++MPG Sbjct: 295 KPGSIKPHDKFEAEVYVLSKEEGGRHTPFTNDYRPQFYFRTTDVTGTIKLPFDKQMVMPG 354 Query: 895 EDSSVKLHLQKKMVLEVGQRFTLR-CGITLGYGVVTKL 1005 ++++ + L K + ++ G +F++R G T+G GVVT++ Sbjct: 355 DNATFSVELIKPIAMQEGLKFSIREGGRTVGAGVVTRI 392
>sp|P42473|EFTU_CHLVI Elongation factor Tu (EF-Tu) Length = 393 Score = 355 bits (912), Expect = 9e-98 Identities = 179/338 (52%), Positives = 241/338 (71%), Gaps = 2/338 (0%) Frame = +1 Query: 1 KRRGITINAAVVDYSTDARHYAHTDCPGHADYIKNMITGANQMEAAILVVAATDGSMPQT 180 + RGITI+ A V+Y TD RHYAH DCPGHADYIKNMITGA QM+ AILVVA TDG MPQT Sbjct: 57 RERGITISTAHVEYQTDKRHYAHIDCPGHADYIKNMITGAAQMDGAILVVAGTDGPMPQT 116 Query: 181 REHLVLAKQIGIDKLVVFINKADAADNEMIELVEMEVRDLLTKYGYDGENIPFVTGSALL 360 REH++LA+Q+ + LVVF+NK D AD E++ELVEME+R+LLT+YG+ G++IP + GSAL Sbjct: 117 REHILLARQVNVPALVVFLNKVDIADPELLELVEMELRELLTEYGFPGDDIPIIKGSALN 176 Query: 361 ALEGKNEEIGKNKIIELLNKIDE-IPLPAREIDKPLFLPVEHVLSITGRGTVCTGRVERG 537 AL G E G+ I+EL++ +D+ IP P R++DKP +PVE V SI+GRGTV TGR+ERG Sbjct: 177 ALNGDPE--GEKAIMELMDAVDDYIPEPVRDVDKPFLMPVEDVFSISGRGTVGTGRIERG 234 Query: 538 VINLGLAVEIIGYNASVKTTITGIEMFRQLLDKAVPGDQVGVLLRSVKKEELRRGMVIIS 717 +I +G VEI+G + K+ +TGIEMF++ LD+ GD G+LLR V KE L RGMVI Sbjct: 235 IIKVGNEVEIVGIKPTTKSVVTGIEMFQKTLDEGQAGDNAGLLLRGVDKEALERGMVIAK 294 Query: 718 PKSVQMYNYVEAQIYLLTEAEGGRKKPSTKNFQSQIYSKSWDCPAFTILPEGKELIMPGE 897 P S+ + +A++Y+L + EGGR P ++ Q Y ++ D LPEG E++MPG+ Sbjct: 295 PGSITPHTKFKAEVYILKKEEGGRHTPFFNGYRPQFYFRTTDVTGSVTLPEGVEMVMPGD 354 Query: 898 DSSVKLHLQKKMVLEVGQRFTLR-CGITLGYGVVTKLL 1008 + SV + L + +E RF +R G T+G G VTK++ Sbjct: 355 NLSVDVELIAPIAMEESLRFAIREGGRTVGAGSVTKIV 392
>sp|P42480|EFTU_TAXOC Elongation factor Tu (EF-Tu) Length = 395 Score = 355 bits (911), Expect = 1e-97 Identities = 182/340 (53%), Positives = 248/340 (72%), Gaps = 4/340 (1%) Frame = +1 Query: 1 KRRGITINAAVVDYSTDARHYAHTDCPGHADYIKNMITGANQMEAAILVVAATDGSMPQT 180 K RGITIN A V+YST RHYAH DCPGHADY+KNM+TGA QM+ AILVVAATDG MPQT Sbjct: 57 KERGITINTAHVEYSTANRHYAHVDCPGHADYVKNMVTGAAQMDGAILVVAATDGPMPQT 116 Query: 181 REHLVLAKQIGIDKLVVFINKADAADN-EMIELVEMEVRDLLTKYGYDGENIPFVTGSAL 357 REH++LA+Q+G+ +LVVF+NK D D+ E++ELVEME+R+LL+ Y +DG+NIP V GSAL Sbjct: 117 REHILLARQVGVPQLVVFMNKVDMVDDPELLELVEMEIRELLSFYDFDGDNIPVVQGSAL 176 Query: 358 LALEGKNEEIGKNKIIELLNKIDE-IPLPAREIDKPLFLPVEHVLSITGRGTVCTGRVER 534 L G + +G I +L++ +D IP+P R D+P +PVE V SITGRGTV TGR+ER Sbjct: 177 GGLNGDAKWVG--TIEQLMDSVDNWIPIPPRLTDQPFLMPVEDVFSITGRGTVATGRIER 234 Query: 535 GVINLGLAVEIIGYNA-SVKTTITGIEMFRQLLDKAVPGDQVGVLLRSVKKEELRRGMVI 711 GVIN G VEI+G A ++K+T+TG+EMFR++LD+ GD VG+LLR ++KE +RRGMVI Sbjct: 235 GVINSGEPVEILGMGAENLKSTVTGVEMFRKILDRGEAGDNVGLLLRGIEKEAIRRGMVI 294 Query: 712 ISPKSVQMYNYVEAQIYLLTEAEGGRKKPSTKNFQSQIYSKSWDCPAFTILPEGKELIMP 891 P SV + +A++Y+L++ EGGR P N++ Q Y ++ D L EG E++MP Sbjct: 295 CKPGSVTPHKKFKAEVYVLSKEEGGRHTPFFNNYRPQFYFRTTDVTGIISLAEGVEMVMP 354 Query: 892 GEDSSVKLHLQKKMVLEVGQRFTLR-CGITLGYGVVTKLL 1008 G++ ++ + L + +E G RF +R G T+G G VT++L Sbjct: 355 GDNVTISVELINAVAMEKGLRFAIREGGRTVGAGQVTEIL 394
>sp|Q9Y700|EFTU_SCHPO Elongation factor Tu, mitochondrial precursor Length = 439 Score = 355 bits (911), Expect = 1e-97 Identities = 178/341 (52%), Positives = 247/341 (72%), Gaps = 5/341 (1%) Frame = +1 Query: 1 KRRGITINAAVVDYSTDARHYAHTDCPGHADYIKNMITGANQMEAAILVVAATDGSMPQT 180 K RGITI++A V+Y T RHYAH DCPGHADYIKNMITGA M+ AI+VV+ATDG MPQT Sbjct: 98 KARGITISSAHVEYETANRHYAHVDCPGHADYIKNMITGAATMDGAIIVVSATDGQMPQT 157 Query: 181 REHLVLAKQIGIDKLVVFINKADAADNEMIELVEMEVRDLLTKYGYDGENIPFVTGSALL 360 REHL+LA+Q+G+ ++VV+INK D + +MIELVEME+R+LL++YG+DG+N P V+GSAL Sbjct: 158 REHLLLARQVGVKQIVVYINKVDMVEPDMIELVEMEMRELLSEYGFDGDNTPIVSGSALC 217 Query: 361 ALEGKNEEIGKNKIIELLNKIDE-IPLPAREIDKPLFLPVEHVLSITGRGTVCTGRVERG 537 ALEG+ EIG N I +L+ +D I LP R+ D P + +E V SI+GRGTV TGRVERG Sbjct: 218 ALEGREPEIGLNSITKLMEAVDSYITLPERKTDVPFLMAIEDVFSISGRGTVVTGRVERG 277 Query: 538 VINLGLAVEIIGYNASVKTTITGIEMFRQLLDKAVPGDQVGVLLRSVKKEELRRGMVIIS 717 + G +EI+GY + +KTT+TGIEMF++ LD AV GD G+LLRS+K+E+L+RGM++ Sbjct: 278 TLKKGAEIEIVGYGSHLKTTVTGIEMFKKQLDAAVAGDNCGLLLRSIKREQLKRGMIVAQ 337 Query: 718 PKSVQMYNYVEAQIYLLTEAEGGRKKPSTKNFQSQIYSKSWDCPAFTILP---EGKELIM 888 P +V + +A Y+LT+ EGGR+ ++ Q+YS++ D P + +++M Sbjct: 338 PGTVAPHQKFKASFYILTKEEGGRRTGFVDKYRPQLYSRTSDVTVELTHPDPNDSDKMVM 397 Query: 889 PGEDSSVKLHLQKKMVLEVGQRFTLR-CGITLGYGVVTKLL 1008 PG++ + L +V+E GQRFT+R G T+G +VT+LL Sbjct: 398 PGDNVEMICTLIHPIVIEKGQRFTVREGGSTVGTALVTELL 438
>sp|O50306|EFTU_BACST Elongation factor Tu (EF-Tu) Length = 395 Score = 355 bits (911), Expect = 1e-97 Identities = 178/340 (52%), Positives = 247/340 (72%), Gaps = 4/340 (1%) Frame = +1 Query: 1 KRRGITINAAVVDYSTDARHYAHTDCPGHADYIKNMITGANQMEAAILVVAATDGSMPQT 180 + RGITI+ A V+Y T+ARHYAH DCPGHADY+KNMITGA QM+ AILVV+A DG MPQT Sbjct: 57 RERGITISTAHVEYETEARHYAHVDCPGHADYVKNMITGAAQMDGAILVVSAADGPMPQT 116 Query: 181 REHLVLAKQIGIDKLVVFINKADAADN-EMIELVEMEVRDLLTKYGYDGENIPFVTGSAL 357 REH++L++Q+G+ +VVF+NK D D+ E++ELVEMEVRDLL++Y + G+ +P + GSAL Sbjct: 117 REHILLSRQVGVPYIVVFLNKCDMVDDEELLELVEMEVRDLLSEYDFPGDEVPVIKGSAL 176 Query: 358 LALEGKNEEIGKNKIIELLNKIDE-IPLPAREIDKPLFLPVEHVLSITGRGTVCTGRVER 534 ALEG + + KIIEL+N +DE IP P RE+DKP +P+E V SITGRGTV TGRVER Sbjct: 177 KALEG--DPKWEEKIIELMNAVDEYIPTPQREVDKPFMMPIEDVFSITGRGTVATGRVER 234 Query: 535 GVINLGLAVEIIGYNASVK-TTITGIEMFRQLLDKAVPGDQVGVLLRSVKKEELRRGMVI 711 G + +G VEIIG + K TT+TG+EMFR+LLD+A GD +G LLR V ++E+ RG V+ Sbjct: 235 GTLKVGDPVEIIGLSDEPKATTVTGVEMFRKLLDQAEAGDNIGALLRGVSRDEVERGQVL 294 Query: 712 ISPKSVQMYNYVEAQIYLLTEAEGGRKKPSTKNFQSQIYSKSWDCPAFTILPEGKELIMP 891 P S+ + +AQ+Y+LT+ EGGR P N++ Q Y ++ D LPEG E++MP Sbjct: 295 AKPGSITPHTKFKAQVYVLTKEEGGRHTPFFSNYRPQFYFRTTDVTGIITLPEGVEMVMP 354 Query: 892 GEDSSVKLHLQKKMVLEVGQRFTLR-CGITLGYGVVTKLL 1008 G++ + + L + +E G +F++R G T+G G V++++ Sbjct: 355 GDNVEMTVELIAPIAIEEGTKFSIREGGRTVGAGSVSEII 394
>sp|Q8KT99|EFTU_RICHE Elongation factor Tu (EF-Tu) Length = 394 Score = 355 bits (910), Expect = 2e-97 Identities = 177/338 (52%), Positives = 246/338 (72%), Gaps = 3/338 (0%) Frame = +1 Query: 1 KRRGITINAAVVDYSTDARHYAHTDCPGHADYIKNMITGANQMEAAILVVAATDGSMPQT 180 K RGITI+ A V+Y T RHYAH DCPGHADY+KNMITGA QM+ AILVV+A DG MPQT Sbjct: 57 KERGITISTAHVEYETKNRHYAHVDCPGHADYVKNMITGAAQMDGAILVVSAADGPMPQT 116 Query: 181 REHLVLAKQIGIDKLVVFINKADAADN-EMIELVEMEVRDLLTKYGYDGENIPFVTGSAL 357 REH++LAKQ+G+ +VVF+NK D D+ +++ELVEMEVR+LL+KYG+ G+ IP + GSAL Sbjct: 117 REHILLAKQVGVPAMVVFLNKVDMVDDPDLLELVEMEVRELLSKYGFPGDEIPVIKGSAL 176 Query: 358 LALEGKNEEIGKNKIIELLNKIDE-IPLPAREIDKPLFLPVEHVLSITGRGTVCTGRVER 534 ALEGK E G+ I EL++ +D IP P R DKP +P+E V SI+GRGTV TGRVE Sbjct: 177 QALEGKPE--GEKAINELMDAVDSYIPQPVRATDKPFLMPIEDVFSISGRGTVVTGRVES 234 Query: 535 GVINLGLAVEIIGYNASVKTTITGIEMFRQLLDKAVPGDQVGVLLRSVKKEELRRGMVII 714 G+I +G +EI+G + KTT TG+EMFR+LLD+ GD VG+LLR K+EE+ RG V+ Sbjct: 235 GIIKVGEEIEIVGLKDTQKTTCTGVEMFRKLLDEGQSGDNVGILLRGTKREEVERGQVLA 294 Query: 715 SPKSVQMYNYVEAQIYLLTEAEGGRKKPSTKNFQSQIYSKSWDCPAFTILPEGKELIMPG 894 P S++ ++ EA++Y+L++ EGGR P T +++ Q Y ++ D LP K+++MPG Sbjct: 295 KPGSIKPHDKFEAEVYVLSKEEGGRHTPFTNDYRPQFYFRTTDVTGTIKLPFDKQMVMPG 354 Query: 895 EDSSVKLHLQKKMVLEVGQRFTLR-CGITLGYGVVTKL 1005 ++++ + L K + ++ G +F++R G T+G GVVTK+ Sbjct: 355 DNATFTVELIKPIAMQEGLKFSIREGGRTVGAGVVTKI 392
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 124,749,189
Number of Sequences: 369166
Number of extensions: 2581236
Number of successful extensions: 9276
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7997
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8346
length of database: 68,354,980
effective HSP length: 112
effective length of database: 47,664,660
effective search space used: 11725506360
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail