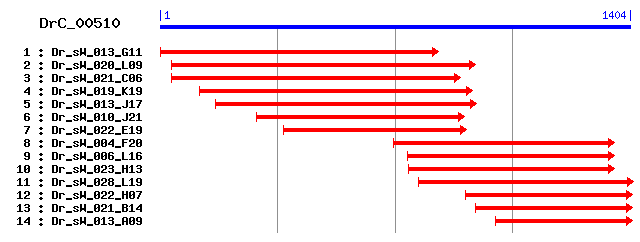
DrC_00510
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00510 (1401 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P68372|TBBX_MOUSE Tubulin beta-? chain >gi|55977480|sp|P... 828 0.0 sp|P11833|TBB_PARLI Tubulin beta chain (Beta tubulin) 827 0.0 sp|P30883|TBB4_XENLA Tubulin beta-4 chain 826 0.0 sp|P02554|TBB_PIG Tubulin beta chain 822 0.0 sp|P09206|TBB3_CHICK Tubulin beta-3 chain (Beta-tubulin cla... 819 0.0 sp|Q7JJU6|TBB2_PANTR Tubulin beta-2 chain >gi|56754798|sp|P... 819 0.0 sp|P32882|TBB2_CHICK Tubulin beta-2 chain (Beta-tubulin cla... 819 0.0 sp|P36221|TBB1_NOTCO Tubulin beta-1 chain (Beta-1 tubulin) 819 0.0 sp|O17449|TBB1_MANSE Tubulin beta-1 chain (Beta-1 tubulin) 819 0.0 sp|Q9YHC3|TBB1_GADMO Tubulin beta-1 chain (Beta-1 tubulin) 819 0.0
>sp|P68372|TBBX_MOUSE Tubulin beta-? chain sp|P68371|TBBX_HUMAN Tubulin beta-? chain (Tubulin beta-2 chain) Length = 445 Score = 828 bits (2138), Expect = 0.0 Identities = 402/430 (93%), Positives = 411/430 (95%) Frame = +1 Query: 22 MREIVHLQAGQCGNQIGSKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEASGGKYV 201 MREIVHLQAGQCGNQIG+KFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEA+GGKYV Sbjct: 1 MREIVHLQAGQCGNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYV 60 Query: 202 PRAVLVDLEPGTMDSVRAGPFGQLFRPDNFIFGQSGAGNNWAKGHYTEGAELVDSVLDVV 381 PRAVLVDLEPGTMDSVR+GPFGQ+FRPDNF+FGQSGAGNNWAKGHYTEGAELVDSVLDVV Sbjct: 61 PRAVLVDLEPGTMDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVV 120 Query: 382 RKEAESCDCLQGFQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVV 561 RKEAESCDCLQGFQ KIREEYPDRIMNTFSVVPSPKVSDTVV Sbjct: 121 RKEAESCDCLQGFQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVV 180 Query: 562 EPYNATLSVHQLVENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 741 EPYNATLSVHQLVENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL Sbjct: 181 EPYNATLSVHQLVENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 240 Query: 742 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM 921 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM Sbjct: 241 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM 300 Query: 922 AACDPRHGRYLTVAAMFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNIKTAVCDIPPRG 1101 AACDPRHGRYLTVAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNN+KTAVCDIPPRG Sbjct: 301 AACDPRHGRYLTVAAVFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRG 360 Query: 1102 LKMSATFIGNSTAIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 1281 LKMSATFIGNSTAIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS Sbjct: 361 LKMSATFIGNSTAIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420 Query: 1282 EYQQYQDATA 1311 EYQQYQDATA Sbjct: 421 EYQQYQDATA 430
>sp|P11833|TBB_PARLI Tubulin beta chain (Beta tubulin) Length = 447 Score = 827 bits (2136), Expect = 0.0 Identities = 401/430 (93%), Positives = 411/430 (95%) Frame = +1 Query: 22 MREIVHLQAGQCGNQIGSKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEASGGKYV 201 MREIVH+QAGQCGNQIG+KFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEA+GGKYV Sbjct: 1 MREIVHMQAGQCGNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYV 60 Query: 202 PRAVLVDLEPGTMDSVRAGPFGQLFRPDNFIFGQSGAGNNWAKGHYTEGAELVDSVLDVV 381 PRAVLVDLEPGTMDSVR+GPFGQ+FRPDNF+FGQSGAGNNWAKGHYTEGAELVDSVLDVV Sbjct: 61 PRAVLVDLEPGTMDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVV 120 Query: 382 RKEAESCDCLQGFQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVV 561 RKEAESCDCLQGFQ KIREEYPDRIMNTFSVVPSPKVSDTVV Sbjct: 121 RKEAESCDCLQGFQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVV 180 Query: 562 EPYNATLSVHQLVENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 741 EPYNATLSVHQLVENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL Sbjct: 181 EPYNATLSVHQLVENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 240 Query: 742 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM 921 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM Sbjct: 241 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM 300 Query: 922 AACDPRHGRYLTVAAMFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNIKTAVCDIPPRG 1101 AACDPRHGRYLTVAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNN+KTAVCDIPPRG Sbjct: 301 AACDPRHGRYLTVAAIFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRG 360 Query: 1102 LKMSATFIGNSTAIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 1281 LKMSATFIGNSTAIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS Sbjct: 361 LKMSATFIGNSTAIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420 Query: 1282 EYQQYQDATA 1311 EYQQYQDATA Sbjct: 421 EYQQYQDATA 430
>sp|P30883|TBB4_XENLA Tubulin beta-4 chain Length = 445 Score = 826 bits (2133), Expect = 0.0 Identities = 401/430 (93%), Positives = 410/430 (95%) Frame = +1 Query: 22 MREIVHLQAGQCGNQIGSKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEASGGKYV 201 MREIVHLQAGQCGNQIG+KFWEVISDEHGIDPTG YHGDSDLQLERINVYYNEA+GGKYV Sbjct: 1 MREIVHLQAGQCGNQIGAKFWEVISDEHGIDPTGAYHGDSDLQLERINVYYNEATGGKYV 60 Query: 202 PRAVLVDLEPGTMDSVRAGPFGQLFRPDNFIFGQSGAGNNWAKGHYTEGAELVDSVLDVV 381 PRAVLVDLEPGTMDSVR+GPFGQ+FRPDNF+FGQSGAGNNWAKGHYTEGAELVDSVLDVV Sbjct: 61 PRAVLVDLEPGTMDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVV 120 Query: 382 RKEAESCDCLQGFQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVV 561 RKEAESCDCLQGFQ KIREEYPDRIMNTFSVVPSPKVSDTVV Sbjct: 121 RKEAESCDCLQGFQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVV 180 Query: 562 EPYNATLSVHQLVENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 741 EPYNATLSVHQLVENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL Sbjct: 181 EPYNATLSVHQLVENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 240 Query: 742 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM 921 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM Sbjct: 241 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM 300 Query: 922 AACDPRHGRYLTVAAMFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNIKTAVCDIPPRG 1101 AACDPRHGRYLTVAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNN+KTAVCDIPPRG Sbjct: 301 AACDPRHGRYLTVAAIFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRG 360 Query: 1102 LKMSATFIGNSTAIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 1281 LKMSATFIGNSTAIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS Sbjct: 361 LKMSATFIGNSTAIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420 Query: 1282 EYQQYQDATA 1311 EYQQYQDATA Sbjct: 421 EYQQYQDATA 430
>sp|P02554|TBB_PIG Tubulin beta chain Length = 445 Score = 822 bits (2122), Expect = 0.0 Identities = 397/430 (92%), Positives = 410/430 (95%) Frame = +1 Query: 22 MREIVHLQAGQCGNQIGSKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEASGGKYV 201 MREIVH+QAGQCGNQIG+KFWEVISDEHGIDPTG+YHGDSDLQLERINVYYNEA+G KYV Sbjct: 1 MREIVHIQAGQCGNQIGAKFWEVISDEHGIDPTGSYHGDSDLQLERINVYYNEAAGNKYV 60 Query: 202 PRAVLVDLEPGTMDSVRAGPFGQLFRPDNFIFGQSGAGNNWAKGHYTEGAELVDSVLDVV 381 PRA+LVDLEPGTMDSVR+GPFGQ+FRPDNF+FGQSGAGNNWAKGHYTEGAELVDSVLDVV Sbjct: 61 PRAILVDLEPGTMDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVV 120 Query: 382 RKEAESCDCLQGFQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVV 561 RKE+ESCDCLQGFQ KIREEYPDRIMNTFSVVPSPKVSDTVV Sbjct: 121 RKESESCDCLQGFQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVV 180 Query: 562 EPYNATLSVHQLVENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 741 EPYNATLSVHQLVENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL Sbjct: 181 EPYNATLSVHQLVENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 240 Query: 742 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM 921 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM Sbjct: 241 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM 300 Query: 922 AACDPRHGRYLTVAAMFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNIKTAVCDIPPRG 1101 AACDPRHGRYLTVAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNN+KTAVCDIPPRG Sbjct: 301 AACDPRHGRYLTVAAVFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRG 360 Query: 1102 LKMSATFIGNSTAIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 1281 LKMSATFIGNSTAIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS Sbjct: 361 LKMSATFIGNSTAIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420 Query: 1282 EYQQYQDATA 1311 EYQQYQDATA Sbjct: 421 EYQQYQDATA 430
>sp|P09206|TBB3_CHICK Tubulin beta-3 chain (Beta-tubulin class-IV) Length = 445 Score = 819 bits (2116), Expect = 0.0 Identities = 398/430 (92%), Positives = 409/430 (95%) Frame = +1 Query: 22 MREIVHLQAGQCGNQIGSKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEASGGKYV 201 MREIVHLQAGQCGNQIG+KFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEA+GGKYV Sbjct: 1 MREIVHLQAGQCGNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYV 60 Query: 202 PRAVLVDLEPGTMDSVRAGPFGQLFRPDNFIFGQSGAGNNWAKGHYTEGAELVDSVLDVV 381 PRAVLVDLEPGTMDSVR+GPF Q+FRPDNF+FGQSGAGNNWAKGHYTEGAELVDSVLDVV Sbjct: 61 PRAVLVDLEPGTMDSVRSGPFRQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVV 120 Query: 382 RKEAESCDCLQGFQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVV 561 RKEAESCDCLQGFQ KIREEYPDRIMNTFSVVPSPKVSDTVV Sbjct: 121 RKEAESCDCLQGFQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVV 180 Query: 562 EPYNATLSVHQLVENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 741 EPYNATLSVHQLVENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL Sbjct: 181 EPYNATLSVHQLVENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 240 Query: 742 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM 921 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVP+LTQQMFDAKNMM Sbjct: 241 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPDLTQQMFDAKNMM 300 Query: 922 AACDPRHGRYLTVAAMFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNIKTAVCDIPPRG 1101 AAC+P HGRYLTVAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNN+KTAVCDIPPRG Sbjct: 301 AACEPGHGRYLTVAAVFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRG 360 Query: 1102 LKMSATFIGNSTAIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 1281 LKMSATFIGNSTAIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS Sbjct: 361 LKMSATFIGNSTAIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420 Query: 1282 EYQQYQDATA 1311 EYQQYQDATA Sbjct: 421 EYQQYQDATA 430
>sp|Q7JJU6|TBB2_PANTR Tubulin beta-2 chain sp|P69895|TBB2_MACMU Tubulin beta-2 chain sp|P07437|TBB2_HUMAN Tubulin beta-2 chain sp|P09244|TBB7_CHICK Tubulin beta-7 chain (Tubulin beta 4') sp|P99024|TBB5_MOUSE Tubulin beta-5 chain sp|P69897|TBB5_RAT Tubulin beta-5 chain sp|P69893|TBB1_CRIGR Tubulin beta-1 chain (Beta-tubulin isotype I) (Class I beta tubulin) Length = 444 Score = 819 bits (2116), Expect = 0.0 Identities = 395/430 (91%), Positives = 410/430 (95%) Frame = +1 Query: 22 MREIVHLQAGQCGNQIGSKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEASGGKYV 201 MREIVH+QAGQCGNQIG+KFWEVISDEHGIDPTGTYHGDSDLQL+RI+VYYNEA+GGKYV Sbjct: 1 MREIVHIQAGQCGNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLDRISVYYNEATGGKYV 60 Query: 202 PRAVLVDLEPGTMDSVRAGPFGQLFRPDNFIFGQSGAGNNWAKGHYTEGAELVDSVLDVV 381 PRA+LVDLEPGTMDSVR+GPFGQ+FRPDNF+FGQSGAGNNWAKGHYTEGAELVDSVLDVV Sbjct: 61 PRAILVDLEPGTMDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVV 120 Query: 382 RKEAESCDCLQGFQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVV 561 RKEAESCDCLQGFQ KIREEYPDRIMNTFSVVPSPKVSDTVV Sbjct: 121 RKEAESCDCLQGFQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVV 180 Query: 562 EPYNATLSVHQLVENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 741 EPYNATLSVHQLVENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL Sbjct: 181 EPYNATLSVHQLVENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 240 Query: 742 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM 921 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQ+FDAKNMM Sbjct: 241 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQVFDAKNMM 300 Query: 922 AACDPRHGRYLTVAAMFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNIKTAVCDIPPRG 1101 AACDPRHGRYLTVAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNN+KTAVCDIPPRG Sbjct: 301 AACDPRHGRYLTVAAVFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRG 360 Query: 1102 LKMSATFIGNSTAIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 1281 LKM+ TFIGNSTAIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS Sbjct: 361 LKMAVTFIGNSTAIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420 Query: 1282 EYQQYQDATA 1311 EYQQYQDATA Sbjct: 421 EYQQYQDATA 430
>sp|P32882|TBB2_CHICK Tubulin beta-2 chain (Beta-tubulin class-II) Length = 445 Score = 819 bits (2116), Expect = 0.0 Identities = 395/430 (91%), Positives = 410/430 (95%) Frame = +1 Query: 22 MREIVHLQAGQCGNQIGSKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEASGGKYV 201 MREIVH+QAGQCGNQIG+KFWEVISDEHGIDPTG+YHGDSDLQLERINVYYNEA+G KYV Sbjct: 1 MREIVHIQAGQCGNQIGAKFWEVISDEHGIDPTGSYHGDSDLQLERINVYYNEATGNKYV 60 Query: 202 PRAVLVDLEPGTMDSVRAGPFGQLFRPDNFIFGQSGAGNNWAKGHYTEGAELVDSVLDVV 381 PRA+LVDLEPGTMDSVR+GPFGQ+FRPDNF+FGQSGAGNNWAKGHYTEGAELVDSVLDVV Sbjct: 61 PRAILVDLEPGTMDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVV 120 Query: 382 RKEAESCDCLQGFQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVV 561 RKE+ESCDCLQGFQ KIREEYPDRIMNTFSV+PSPKVSDTVV Sbjct: 121 RKESESCDCLQGFQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVMPSPKVSDTVV 180 Query: 562 EPYNATLSVHQLVENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 741 EPYNATLSVHQLVENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL Sbjct: 181 EPYNATLSVHQLVENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 240 Query: 742 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM 921 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFD+KNMM Sbjct: 241 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDSKNMM 300 Query: 922 AACDPRHGRYLTVAAMFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNIKTAVCDIPPRG 1101 AACDPRHGRYLTVAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNN+KTAVCDIPPRG Sbjct: 301 AACDPRHGRYLTVAAIFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRG 360 Query: 1102 LKMSATFIGNSTAIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 1281 LKMSATFIGNSTAIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS Sbjct: 361 LKMSATFIGNSTAIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420 Query: 1282 EYQQYQDATA 1311 EYQQYQDATA Sbjct: 421 EYQQYQDATA 430
>sp|P36221|TBB1_NOTCO Tubulin beta-1 chain (Beta-1 tubulin) Length = 446 Score = 819 bits (2115), Expect = 0.0 Identities = 396/430 (92%), Positives = 408/430 (94%) Frame = +1 Query: 22 MREIVHLQAGQCGNQIGSKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEASGGKYV 201 MREIVHLQAGQCGNQIGSKFWEVISDEHGIDPTG+YHGDSDLQL+RINVYYNEASGGKYV Sbjct: 1 MREIVHLQAGQCGNQIGSKFWEVISDEHGIDPTGSYHGDSDLQLDRINVYYNEASGGKYV 60 Query: 202 PRAVLVDLEPGTMDSVRAGPFGQLFRPDNFIFGQSGAGNNWAKGHYTEGAELVDSVLDVV 381 PRAVLVDLEPGTMDSVR+GPFGQ+FRPDNF+FGQSGAGNNWAKGHYTEGAELVDSVLDVV Sbjct: 61 PRAVLVDLEPGTMDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVV 120 Query: 382 RKEAESCDCLQGFQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVV 561 RKEAE CDCLQGFQ KIREEYPDRIMNTFSVVPSPKVSDTVV Sbjct: 121 RKEAEGCDCLQGFQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVV 180 Query: 562 EPYNATLSVHQLVENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 741 EPYNATLSVHQLVENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL Sbjct: 181 EPYNATLSVHQLVENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 240 Query: 742 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM 921 RFPGQLNADLRKLAVNMVPFPRLHFF+PGFAPLTSRG QQYR+LTVPELTQQMFD+KNMM Sbjct: 241 RFPGQLNADLRKLAVNMVPFPRLHFFIPGFAPLTSRGGQQYRSLTVPELTQQMFDSKNMM 300 Query: 922 AACDPRHGRYLTVAAMFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNIKTAVCDIPPRG 1101 AACDPRHGRYLTVAA+FRGRMSMKEVDEQMLN QNKNSSYFVEWIPNN+KTAVCDIPPRG Sbjct: 301 AACDPRHGRYLTVAAIFRGRMSMKEVDEQMLNAQNKNSSYFVEWIPNNVKTAVCDIPPRG 360 Query: 1102 LKMSATFIGNSTAIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 1281 LKM+ATFIGNSTAIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS Sbjct: 361 LKMAATFIGNSTAIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420 Query: 1282 EYQQYQDATA 1311 EYQQYQDATA Sbjct: 421 EYQQYQDATA 430
>sp|O17449|TBB1_MANSE Tubulin beta-1 chain (Beta-1 tubulin) Length = 447 Score = 819 bits (2115), Expect = 0.0 Identities = 394/430 (91%), Positives = 409/430 (95%) Frame = +1 Query: 22 MREIVHLQAGQCGNQIGSKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEASGGKYV 201 MREIVH+QAGQCGNQIG+KFWE+ISDEHGIDPTG YHGDSDLQLERINVYYNEASGGKYV Sbjct: 1 MREIVHIQAGQCGNQIGAKFWEIISDEHGIDPTGAYHGDSDLQLERINVYYNEASGGKYV 60 Query: 202 PRAVLVDLEPGTMDSVRAGPFGQLFRPDNFIFGQSGAGNNWAKGHYTEGAELVDSVLDVV 381 PRA+LVDLEPGTMDSVR+GPFGQ+FRPDNF+FGQSGAGNNWAKGHYTEGAELVDSVLDVV Sbjct: 61 PRAILVDLEPGTMDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVV 120 Query: 382 RKEAESCDCLQGFQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVV 561 RKEAESCDCLQGFQ KIREEYPDRIMNT+SVVPSPKVSDTVV Sbjct: 121 RKEAESCDCLQGFQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTYSVVPSPKVSDTVV 180 Query: 562 EPYNATLSVHQLVENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 741 EPYNATLSVHQLVENTDET+CIDNEALYDICFRTLKL+TPTYGDLNHLVS TMSGVTTCL Sbjct: 181 EPYNATLSVHQLVENTDETYCIDNEALYDICFRTLKLSTPTYGDLNHLVSLTMSGVTTCL 240 Query: 742 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM 921 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM Sbjct: 241 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM 300 Query: 922 AACDPRHGRYLTVAAMFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNIKTAVCDIPPRG 1101 AACDPRHGRYLTVAA+FRGRMSMKEVDEQMLN+QNKNSSYFVEWIPNN+KTAVCDIPPRG Sbjct: 301 AACDPRHGRYLTVAAIFRGRMSMKEVDEQMLNIQNKNSSYFVEWIPNNVKTAVCDIPPRG 360 Query: 1102 LKMSATFIGNSTAIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 1281 LKMSATFIGNSTAIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS Sbjct: 361 LKMSATFIGNSTAIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420 Query: 1282 EYQQYQDATA 1311 EYQQYQ+ATA Sbjct: 421 EYQQYQEATA 430
>sp|Q9YHC3|TBB1_GADMO Tubulin beta-1 chain (Beta-1 tubulin) Length = 445 Score = 819 bits (2115), Expect = 0.0 Identities = 397/430 (92%), Positives = 409/430 (95%) Frame = +1 Query: 22 MREIVHLQAGQCGNQIGSKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEASGGKYV 201 MREIVHLQAGQCGNQIG+KFWEVISDEHGIDPTGTYHGDSDLQL+RINVYYNEASGGKYV Sbjct: 1 MREIVHLQAGQCGNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLDRINVYYNEASGGKYV 60 Query: 202 PRAVLVDLEPGTMDSVRAGPFGQLFRPDNFIFGQSGAGNNWAKGHYTEGAELVDSVLDVV 381 PRAVLVDLEPGTMDSVR+G FGQ+FRPDNF+FGQSGAGNNWAKGHYTEGAELVDSVLDVV Sbjct: 61 PRAVLVDLEPGTMDSVRSGAFGQVFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVV 120 Query: 382 RKEAESCDCLQGFQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVV 561 RKEAESCDCLQGFQ KIREEYPDRIMNTFSVVPSPKVSDTVV Sbjct: 121 RKEAESCDCLQGFQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVV 180 Query: 562 EPYNATLSVHQLVENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCL 741 EPYNATLSVHQLVENTDET+CIDNEALYDICFRTLKLTTP+YGDLNHLVSATMSGVTTCL Sbjct: 181 EPYNATLSVHQLVENTDETYCIDNEALYDICFRTLKLTTPSYGDLNHLVSATMSGVTTCL 240 Query: 742 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMM 921 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYR+LTVPELTQQMFD KNMM Sbjct: 241 RFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRSLTVPELTQQMFDGKNMM 300 Query: 922 AACDPRHGRYLTVAAMFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNIKTAVCDIPPRG 1101 AACDPRHGRYLTVAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNN+KTAVCDIPPRG Sbjct: 301 AACDPRHGRYLTVAAIFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRG 360 Query: 1102 LKMSATFIGNSTAIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 1281 LKM+ATFIGNSTAIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS Sbjct: 361 LKMAATFIGNSTAIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420 Query: 1282 EYQQYQDATA 1311 EYQQYQDATA Sbjct: 421 EYQQYQDATA 430
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 158,613,604
Number of Sequences: 369166
Number of extensions: 3281833
Number of successful extensions: 9371
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8115
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8647
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 16647906880
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail