DrC_00477
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
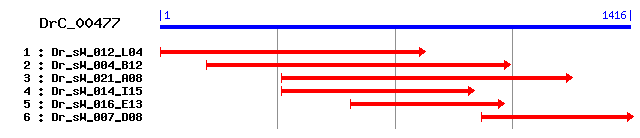
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00477 (1414 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q96T60|PNKP_HUMAN Bifunctional polynucleotide phosphatas... 280 7e-75 sp|Q9JLV6|PNKP_MOUSE Bifunctional polynucleotide phosphatas... 270 6e-72 sp|Q19683|YZR5_CAEEL Hypothetical protein F21D5.5 in chromo... 269 1e-71 sp|O13911|PNK1_SCHPO Bifunctional polynucleotide phosphatas... 156 1e-37 sp|P21286|Y033_NPVAC Hypothetical 20.8 kDa protein in FGF-V... 107 1e-22 sp|Q03796|TPP1_YEAST Polynucleotide 3'-phosphatase (2'(3')-... 57 2e-07 sp|Q9HET9|TPP1_SACMI Polynucleotide 3'-phosphatase (2'(3')-... 50 2e-05 sp|Q87QK9|HIS7_VIBPA Histidine biosynthesis bifunctional pr... 43 0.003 sp|Q9KSX1|HIS7_VIBCH Histidine biosynthesis bifunctional pr... 41 0.010 sp|P08116|PVA_PLAFA Processed variable antigen 40 0.013
>sp|Q96T60|PNKP_HUMAN Bifunctional polynucleotide phosphatase/kinase (Polynucleotide kinase-3'-phosphatase) (DNA 5'-kinase/3'-phosphatase) [Includes: Polynucleotide 3'-phosphatase (2'(3')-polynucleotidase); Polynucleotide 5'-hydroxyl-kinase ] Length = 521 Score = 280 bits (716), Expect = 7e-75 Identities = 170/413 (41%), Positives = 235/413 (56%), Gaps = 2/413 (0%) Frame = +2 Query: 2 EKSETVESKDDTIHEA-VETNEEKMDKFKRKLGMVKDEEEDKDWVKGNGYLVYTPSDIKS 178 E++ T ES+ DT + + +EK D K M K + W LV+T + +K Sbjct: 107 EETRTPESQPDTPPGTPLVSQDEKRDAELPKKRMRKS---NPGWENLEKLLVFTAAGVKP 163 Query: 179 SEKIASFDLDGTLIDTKSGKKFAEGINDWKIKFNEIPNKLKTLEDEGYKLVIFTNQGGIE 358 K+A FDLDGTLI T+SGK F G +DW+I + EIP KL+ LE EGYKLVIFTNQ I Sbjct: 164 QGKVAGFDLDGTLITTRSGKVFPTGPSDWRILYPEIPRKLRELEAEGYKLVIFTNQMSIG 223 Query: 359 XXXXXXXXXXXXXXAIIEQLAVKIQVLVATNYSNVYRKPNTGMFDLLDKEHNSGIKINCD 538 A++E+L V QVLVAT ++ +YRKP TGM+D L ++ N G I+ Sbjct: 224 RGKLPAEEFKAKVEAVVEKLGVPFQVLVAT-HAGLYRKPVTGMWDHLQEQANDGTPISIG 282 Query: 539 ESLYVGDAAGRVADWAP-KMKKDFSNSDRLFALNCHLKFYTPEAFFLQQEEGEFKMPEFH 715 +S++VGDAAGR A+WAP + KKDFS +DRLFALN L F TPE FFL+ F++P F Sbjct: 283 DSIFVGDAAGRPANWAPGRKKKDFSCADRLFALNLGLPFATPEEFFLKWPAAGFELPAFD 342 Query: 716 PAETVKNCPTLMIKYDKFQKSASDIKDILPKDSSEVVLILMVGYPGSGKSTFCKKYLEPL 895 P TV L + + SAS +++ VG+PG+GKSTF KK+L Sbjct: 343 P-RTVSRSGPLCLPESRALLSASP------------EVVVAVGFPGAGKSTFLKKHLVSA 389 Query: 896 GFNVINRDTLKTWQKCVAETQSLFXXXXXXXXXXTNPDLATRDRYISAAVECGVKHIICC 1075 G+ +NRDTL +WQ+CV ++ TNPD A+R RY+ A GV C Sbjct: 390 GYVHVNRDTLGSWQRCVTTCETALKQGKRVAIDNTNPDAASRARYVQCARAAGVP-CRCF 448 Query: 1076 IMDVSEDQSRHNEHFRQLTADHHDHVSEMVFKIHKNKYVEPSDKEKFTEICKI 1234 + + +Q+RHN FR++T H VS+MV ++ ++ P+ E F+ I +I Sbjct: 449 LFTATLEQARHNNRFREMTDSSHIPVSDMVMYGYRKQFEAPTLAEGFSAILEI 501
>sp|Q9JLV6|PNKP_MOUSE Bifunctional polynucleotide phosphatase/kinase (Polynucleotide kinase-3'-phosphatase) (DNA 5'-kinase/3'-phosphatase) [Includes: Polynucleotide 3'-phosphatase (2'(3')-polynucleotidase); Polynucleotide 5'-hydroxyl-kinase ] Length = 522 Score = 270 bits (691), Expect = 6e-72 Identities = 165/398 (41%), Positives = 231/398 (58%), Gaps = 1/398 (0%) Frame = +2 Query: 44 EAVETNEEKMDKFKRKLGMVKDEEEDKDWVKGNGYLVYTPSDIKSSEKIASFDLDGTLID 223 E +T +K K LG W LV+T S +K K+A+FDLDGTLI Sbjct: 128 EGEDTEPQKKRVRKSSLG----------WESLKKLLVFTASGVKPQGKVAAFDLDGTLIT 177 Query: 224 TKSGKKFAEGINDWKIKFNEIPNKLKTLEDEGYKLVIFTNQGGIEXXXXXXXXXXXXXXA 403 T+SGK F +DW+I + EIP KL+ L EGYKLVIFTNQ GI A Sbjct: 178 TRSGKVFPTSPSDWRILYPEIPKKLQELAAEGYKLVIFTNQMGIGRGKLPAEVFKGKVEA 237 Query: 404 IIEQLAVKIQVLVATNYSNVYRKPNTGMFDLLDKEHNSGIKINCDESLYVGDAAGRVADW 583 ++E+L V QVLVAT ++ + RKP +GM+D L ++ N GI I+ ++S++VGDAAGR+A+W Sbjct: 238 VLEKLGVPFQVLVAT-HAGLNRKPVSGMWDHLQEQANEGIPISVEDSVFVGDAAGRLANW 296 Query: 584 AP-KMKKDFSNSDRLFALNCHLKFYTPEAFFLQQEEGEFKMPEFHPAETVKNCPTLMIKY 760 AP + KKDFS +DRLFALN L F TPE FFL+ F++P F P T+ + L + Sbjct: 297 APGRKKKDFSCADRLFALNVGLPFATPEEFFLKWPAARFELPAFDP-RTISSAGPLYL-- 353 Query: 761 DKFQKSASDIKDILPKDSSEVVLILMVGYPGSGKSTFCKKYLEPLGFNVINRDTLKTWQK 940 +S+S L + EVV + VG+PG+GKSTF +++L G+ +NRDTL +WQ+ Sbjct: 354 ---PESSS-----LLSPNPEVV--VAVGFPGAGKSTFIQEHLVSAGYVHVNRDTLGSWQR 403 Query: 941 CVAETQSLFXXXXXXXXXXTNPDLATRDRYISAAVECGVKHIICCIMDVSEDQSRHNEHF 1120 CV+ Q+ TNPD+ +R RYI A + GV C + +Q+RHN F Sbjct: 404 CVSSCQAALRQGKRVVIDNTNPDVPSRARYIQCAKDAGVP-CRCFNFCATIEQARHNNRF 462 Query: 1121 RQLTADHHDHVSEMVFKIHKNKYVEPSDKEKFTEICKI 1234 R++T H VS+MV ++ ++ P+ E F EI +I Sbjct: 463 REMTDPSHAPVSDMVMFSYRKQFEPPTLAEGFLEILEI 500
>sp|Q19683|YZR5_CAEEL Hypothetical protein F21D5.5 in chromosome IV Length = 407 Score = 269 bits (688), Expect = 1e-71 Identities = 163/393 (41%), Positives = 226/393 (57%), Gaps = 5/393 (1%) Frame = +2 Query: 134 KGNGYL-VYTPSDIKSSEKIASFDLDGTLIDTKSGKKFAEGINDWKIKFNEIPNKLKTLE 310 K NG L ++T S+ + EKIA+FD+DGTLI TKSGK F DW++ ++ IP+ K L Sbjct: 34 KDNGDLMIFTHSECEGKEKIAAFDMDGTLIKTKSGKVFPTNCQDWQLLYDSIPSDFKKLH 93 Query: 311 DEGYKLVIFTNQGGIEXXXXXXXXXXXXXXAIIEQLAVKIQVLVATNYSNVYRKPNTGMF 490 +G+K+VIFTNQ GI AI+ +L + +Q V+ + YRKP GM+ Sbjct: 94 SDGFKIVIFTNQKGIHAGKVDRNEFRKKIEAIVGKLGIPVQAFVSVAGGH-YRKPCVGMW 152 Query: 491 DLLDKEHNSGIKINCDESLYVGDAAGRVADWAPKMKKDFSNSDRLFALNCHLKFYTPEAF 670 + L K N ++IN ES++VGDAAGR+ + KKD S +DR FA N +KF TPE F Sbjct: 153 NEL-KLRNDEVEINEKESIFVGDAAGRIKT-TSRPKKDHSYADRFFAANVGVKFQTPEEF 210 Query: 671 FLQQEEGEFKMPEFHPAETVKNCPTLMIKYDKFQKSASDIKDILPKD----SSEVVLILM 838 F G+ K+ E P +D + +I ++ P D SSE +ILM Sbjct: 211 F-----GKSKVDE----------PWGPPNFDPKNLFSEEITELEPHDAQLKSSEKEIILM 255 Query: 839 VGYPGSGKSTFCKKYLEPLGFNVINRDTLKTWQKCVAETQSLFXXXXXXXXXXTNPDLAT 1018 VG+PGSGKSTF K + ++NRDT+ TWQKCVA T+S T+PDL + Sbjct: 256 VGFPGSGKSTFAKMLGHQHDYKIVNRDTIGTWQKCVAATRSYLADGKSVVIDNTSPDLES 315 Query: 1019 RDRYISAAVECGVKHIICCIMDVSEDQSRHNEHFRQLTADHHDHVSEMVFKIHKNKYVEP 1198 R RYI A E GV I C M+ S + ++HN FR LT D+ +S MV +IHK KYVEP Sbjct: 316 RKRYIDVAKELGVP-IRCFEMNCSMEHAQHNIRFRVLTDDNAAEISSMVLRIHKGKYVEP 374 Query: 1199 SDKEKFTEICKIVPALEFENDDHKCIYSKYLLE 1297 + E F++I K+ +FE ++H+ +Y YL+E Sbjct: 375 TLSEGFSQIVKVNFRPKFEVEEHEKLYKMYLIE 407
>sp|O13911|PNK1_SCHPO Bifunctional polynucleotide phosphatase/kinase (Polynucleotide kinase-3'-phosphatase) (DNA 5'-kinase/3'-phosphatase) [Includes: Polynucleotide 3'-phosphatase (2'(3')-polynucleotidase); Polynucleotide 5'-hydroxy-kinase ] Length = 421 Score = 156 bits (395), Expect = 1e-37 Identities = 125/420 (29%), Positives = 189/420 (45%), Gaps = 26/420 (6%) Frame = +2 Query: 44 EAVETNEEKMDKFKRKLGMVKDEEEDKD----------WVKGNGYLVYTPSDIKSSEKIA 193 E++ + EK K +K G + + W + + +K ++K Sbjct: 12 ESLTSYFEKSSKSSKKYGSQNKDSDSSSTCLQQKIEIQWSITDSLYIAKYGKLKKTKKFI 71 Query: 194 SFDLDGTLIDTKSGKKFAEGINDWKIKFNEIPNKLKTLEDEGYKLVIFTNQGGIEXXXXX 373 +FDLDGTLI TKSG+ F++ DW + KLK L + Y LVIF+NQ GI Sbjct: 72 AFDLDGTLIKTKSGRVFSKDAADWTWWHPSVVPKLKALYQDNYSLVIFSNQNGIPRKPSA 131 Query: 374 XXXXXXXXXAIIEQLAVKIQVLVATNYSNVYRKPNTGMFDLLDKEHNSGIKINCDESLYV 553 AI E L + I VL A + +RKP TGM++ K+ N I ++ + YV Sbjct: 132 GHTFQMKIRAIFESLDLPI-VLYAAILKDKFRKPLTGMWNSFLKDVNRSIDLSFIK--YV 188 Query: 554 GDAAGRVADWAPKMKKDFSNSDRLFALNCHLKFYTPEAFFLQQEEGEFKMPEFHPAE-TV 730 GDAAGR D +++D FA N +KF TPE FFL FHP V Sbjct: 189 GDAAGRPG--------DHNSTDLKFAENIGIKFETPEQFFLGHSFVPPNFESFHPKNYLV 240 Query: 731 KNCPTLMIKYDKFQKSASDIKDILPKDSSEVVLILMVGYPGSGKSTFCKKYLEPLGFNVI 910 +N + + K S ++++VG+P SGKST + + G+ + Sbjct: 241 RNSSSHPYHF---------------KKSEHQEIVVLVGFPSSGKSTLAESQIVTQGYERV 285 Query: 911 NRDTLKTWQKCV-AETQSL------------FXXXXXXXXXXTNPDLATRDRYISAAVEC 1051 N+D LKT KC+ A ++L TNP + +R +I A E Sbjct: 286 NQDILKTKSKCIKAAIEALKKEKSVVIGMYSIISTTYAISDNTNPTIESRKMWIDIAQEF 345 Query: 1052 GVKHIICCIMDVSEDQSRHNEHFRQLTADHHD--HVSEMVFKIHKNKYVEPSDKEKFTEI 1225 + I C + SE+ +RHN FR + HH+ + E+ F K+++ P+ +E FT + Sbjct: 346 EIP-IRCIHLQSSEELARHNNVFRYI---HHNQKQLPEIAFNSFKSRFQMPTVEEGFTNV 401
>sp|P21286|Y033_NPVAC Hypothetical 20.8 kDa protein in FGF-VUBI intergenic region (ORF 1) Length = 182 Score = 107 bits (266), Expect = 1e-22 Identities = 69/172 (40%), Positives = 96/172 (55%) Frame = +2 Query: 155 YTPSDIKSSEKIASFDLDGTLIDTKSGKKFAEGINDWKIKFNEIPNKLKTLEDEGYKLVI 334 Y D KIA+FDLDGTLI +K+ KF + +DW++ +KLK L + GY LV+ Sbjct: 12 YAVHDGAKRTKIAAFDLDGTLISSKTRSKFPKNPDDWQLL--PCAHKLKRLYELGYDLVV 69 Query: 335 FTNQGGIEXXXXXXXXXXXXXXAIIEQLAVKIQVLVATNYSNVYRKPNTGMFDLLDKEHN 514 FTNQ + I + V I V+ N + +RKP+TGM+ + K+ Sbjct: 70 FTNQAHLGSGKIKASDLLYKLENIKKATGVPISFYVSPN-KDEHRKPDTGMWREMAKQFT 128 Query: 515 SGIKINCDESLYVGDAAGRVADWAPKMKKDFSNSDRLFALNCHLKFYTPEAF 670 I+ ++S YVGDAAGR+ +KDFS+SDR+FA N L+FYTPE F Sbjct: 129 ---HIDKEQSFYVGDAAGRIN--LTTGQKDFSDSDRVFAKNLSLQFYTPEQF 175
>sp|Q03796|TPP1_YEAST Polynucleotide 3'-phosphatase (2'(3')-polynucleotidase) (DNA 3' phosphatase) (Three prime phosphatase) Length = 238 Score = 56.6 bits (135), Expect = 2e-07 Identities = 61/214 (28%), Positives = 83/214 (38%), Gaps = 52/214 (24%) Frame = +2 Query: 188 IASFDLDGTLIDTKSGK-KFAEGINDWK-IKFNEIPNKLKTL-----EDEGYKLVIFTNQ 346 + +FDLD T+I KS F+ +DW+ I FN + L L D +VIF+NQ Sbjct: 31 VYAFDLDHTIIKPKSPNISFSRSASDWQFINFNSKKSTLDYLCNIIDNDPTAVIVIFSNQ 90 Query: 347 GGIEXXXXXXXXXXXXXXAIIEQL--------------------AVKIQVLVATNYSNV- 463 GG+ I+ L A K A N+S + Sbjct: 91 GGVITVPRTSKSCTKYTNKILLFLKAIKNDERGETLSHRLWLYAAPKRPKTFAANHSKIT 150 Query: 464 -----------------YRKPNTGMFDLLDKEHNSGIKINCDES-------LYVGDAAGR 571 RKP TGM + ++ S +++ S Y GDAAGR Sbjct: 151 FASLGESYNNDPNIFEKVRKPMTGMVEFFKRDLESAYRVSEQISPIKLNWIYYCGDAAGR 210 Query: 572 VADWAPKMKKDFSNSDRLFALNCHLKFYTPEAFF 673 KKDFS+SD FA N H++F PE F Sbjct: 211 --------KKDFSDSDIKFAENLHVEFKYPEEIF 236
>sp|Q9HET9|TPP1_SACMI Polynucleotide 3'-phosphatase (2'(3')-polynucleotidase) (DNA 3' phosphatase) (Three prime phosphatase) Length = 245 Score = 49.7 bits (117), Expect = 2e-05 Identities = 58/216 (26%), Positives = 82/216 (37%), Gaps = 54/216 (25%) Frame = +2 Query: 188 IASFDLDGTLIDTKS-GKKFAEGINDWKIKFNEIPNKLKTLE--------DEGYKLVIFT 340 + +FDLD T+I KS K++ NDW+ F +K TL+ D +VIF+ Sbjct: 38 VYAFDLDHTIIKPKSPNAKYSRSANDWQ--FMNFDSKKSTLDYLFSITDNDLAAVIVIFS 95 Query: 341 NQGGIEXXXXXXXXXXXXXXAII------------EQLAVKIQVLVAT------------ 448 NQGG+ I E+L+ ++ + A Sbjct: 96 NQGGVITVPRTSKSCSKYINKISLFLKAIENDKRGEKLSPRLWIYAAPKRPKTVVTNNKK 155 Query: 449 --------NYSN------VYRKPNTGMFDLLDKEHNSGIK-------INCDESLYVGDAA 565 +Y+N RKP TGM D + + I + Y GDAA Sbjct: 156 ITFPSLCKSYNNDPEIFEKVRKPMTGMADFFRIDITDACRVLKTVPPIKLNWVYYCGDAA 215 Query: 566 GRVADWAPKMKKDFSNSDRLFALNCHLKFYTPEAFF 673 GR K DFS+SD FA H++F PE F Sbjct: 216 GR--------KNDFSDSDIKFAEKLHVEFKYPEEVF 243
>sp|Q87QK9|HIS7_VIBPA Histidine biosynthesis bifunctional protein hisB [Includes: Histidinol-phosphatase ; Imidazoleglycerol-phosphate dehydratase (IGPD)] Length = 357 Score = 42.7 bits (99), Expect = 0.003 Identities = 40/137 (29%), Positives = 61/137 (44%), Gaps = 7/137 (5%) Frame = +2 Query: 170 IKSSEKIASFDLDGTLIDTKSGKKFAEGINDWKIKFNEIPNKLKTLEDEGYKLVIFTNQG 349 + +KI D DGTLI + ++ K++ IP+ L +L+D GY+LV+ TNQ Sbjct: 1 MSKQQKILFIDRDGTLIVEPPIDFQVDRLDKLKLEPFVIPSLL-SLQDAGYRLVMVTNQD 59 Query: 350 GI---EXXXXXXXXXXXXXXAIIEQLAVKI-QVLVATNYSN---VYRKPNTGMFDLLDKE 508 G+ I E VK VL+ ++ RKP GM KE Sbjct: 60 GLGTDSYPQEDFDAPHNMMMEIFESQGVKFDDVLICPHFEEDNCSCRKPKLGMV----KE 115 Query: 509 HNSGIKINCDESLYVGD 559 + G K++ S+ +GD Sbjct: 116 YLQGGKVDFQNSVVIGD 132
>sp|Q9KSX1|HIS7_VIBCH Histidine biosynthesis bifunctional protein hisB [Includes: Histidinol-phosphatase ; Imidazoleglycerol-phosphate dehydratase (IGPD)] Length = 357 Score = 40.8 bits (94), Expect = 0.010 Identities = 40/137 (29%), Positives = 60/137 (43%), Gaps = 7/137 (5%) Frame = +2 Query: 170 IKSSEKIASFDLDGTLIDTKSGKKFAEGINDWKIKFNEIPNKLKTLEDEGYKLVIFTNQG 349 + +KI D DGTLI + ++ K++ IP+ LK L++ GY+LV+ TNQ Sbjct: 1 MSKQQKILFIDRDGTLIVEPPVDFQVDRLDKLKLEPYVIPSLLK-LQEAGYRLVMVTNQD 59 Query: 350 GI---EXXXXXXXXXXXXXXAIIEQLAVKI-QVLVATNYSN---VYRKPNTGMFDLLDKE 508 G+ I E VK VL+ ++ RKP G L KE Sbjct: 60 GLGTSSYPQADFDAPHNMMMDIFESQGVKFNDVLICPHFERDNCSCRKPKLG----LVKE 115 Query: 509 HNSGIKINCDESLYVGD 559 + G K++ S +GD Sbjct: 116 YLQGGKVDFKSSAVIGD 132
>sp|P08116|PVA_PLAFA Processed variable antigen Length = 224 Score = 40.4 bits (93), Expect = 0.013 Identities = 35/111 (31%), Positives = 49/111 (44%), Gaps = 3/111 (2%) Frame = +2 Query: 2 EKSETVESKDDTIHEAVETNEEKMDKFKRKLGMVKDEEEDKDWVKGNGYLVYTPSDIKSS 181 E ET ESK+ I+E ET K+ R+ VK EE KD G V P + + Sbjct: 94 ESKETGESKETRIYE--ETKYNKITSEFRETENVKITEESKD---REGNKVSGPYENSEN 148 Query: 182 EKIASFDLDGTLIDTKS---GKKFAEGINDWKIKFNEIPNKLKTLEDEGYK 325 + S + + K G+K E +ND + +E P KL E+ G K Sbjct: 149 SNVTSESEETKKLAEKEENEGEKLGENVNDGASENSEDPKKLTEQEENGTK 199
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 152,639,880
Number of Sequences: 369166
Number of extensions: 3129233
Number of successful extensions: 10328
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9735
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10293
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 16837087640
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail