DrC_00397
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
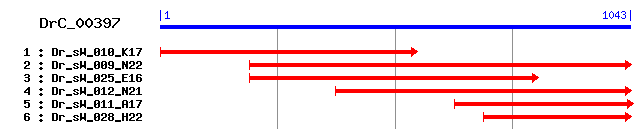
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00397 (1043 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q96EY1|DNJA3_HUMAN DnaJ homolog subfamily A member 3, mi... 242 1e-63 sp|Q99M87|DNJA3_MOUSE DnaJ homolog subfamily A member 3, mi... 240 4e-63 sp|Q24331|TID_DROVI Tumorous imaginal discs protein, mitoch... 236 6e-62 sp|Q27237|TID_DROME Tumorous imaginal discs protein, mitoch... 225 2e-58 sp|Q8TA83|DNJ10_CAEEL DnaJ homolog dnj-10 (DnaJ domain prot... 179 2e-44 sp|O69269|DNAJ_BACSH Chaperone protein dnaJ 161 3e-39 sp|Q892R1|DNAJ_CLOTE Chaperone protein dnaJ 161 3e-39 sp|Q5KWZ8|DNAJ_GEOKA Chaperone protein dnaJ 160 6e-39 sp|P63969|DNAJ_NEIMB Chaperone protein dnaJ >gi|54040948|sp... 159 2e-38 sp|Q5F5M1|DNAJ_NEIG1 Chaperone protein dnaJ 157 4e-38
>sp|Q96EY1|DNJA3_HUMAN DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1) Length = 480 Score = 242 bits (617), Expect = 1e-63 Identities = 116/252 (46%), Positives = 171/252 (67%), Gaps = 1/252 (0%) Frame = +3 Query: 75 ERAQEVNVNLTFGQAACGVNKELYVNLMMECGRCGGSKSEPGTSSTVCPSCQGSGVQSVN 254 ++ QE + LTF QAA GVNKE VN+M C RC G +EPGT C C GSG++++N Sbjct: 206 DQPQEYFMELTFNQAAKGVNKEFTVNIMDTCERCNGKGNEPGTKVQHCHYCGGSGMETIN 265 Query: 255 TGPFLMXXXXXXXXXXXXIIKNPCHDCKGKGRVSQRKKIRVSIPAGIEDGQSLRVSLGAR 434 TGPF+M II +PC C+G G+ Q+K++ + +PAG+EDGQ++R+ +G R Sbjct: 266 TGPFVMRSTCRRCGGRGSIIISPCVVCRGAGQAKQKKRVMIPVPAGVEDGQTVRMPVGKR 325 Query: 435 SAQELYVTVRVEKSNLFRRDGSDVHSDITVSLPLAALGGKITVPGIYGDVTVDIPKGSCS 614 E+++T RV+KS +FRRDG+D+HSD+ +S+ A LGG G+Y + V IP G+ + Sbjct: 326 ---EIFITFRVQKSPVFRRDGADIHSDLFISIAQALLGGTARAQGLYETINVTIPPGTQT 382 Query: 615 HDRIRLPGKGIARVNSYGYGDHYLNIKIQPTKRLTETQRALLLALAEQD-ESAGMINGVT 791 +IR+ GKGI R+NSYGYGDHY++IKI+ KRLT Q++L+L+ AE + + G +NGVT Sbjct: 383 DQKIRMGGKGIPRINSYGYGDHYIHIKIRVPKRLTSRQQSLILSYAEDETDVEGTVNGVT 442 Query: 792 MTANGKQAIDDT 827 +T++G +D + Sbjct: 443 LTSSGGSTMDSS 454
>sp|Q99M87|DNJA3_MOUSE DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (mTid-1) Length = 480 Score = 240 bits (613), Expect = 4e-63 Identities = 118/252 (46%), Positives = 168/252 (66%), Gaps = 1/252 (0%) Frame = +3 Query: 75 ERAQEVNVNLTFGQAACGVNKELYVNLMMECGRCGGSKSEPGTSSTVCPSCQGSGVQSVN 254 ++ QE + LTF QAA GVNKE VN+M C RC G +EPGT C C GSG++++N Sbjct: 206 DQPQEYIMELTFNQAAKGVNKEFTVNIMDTCERCDGKGNEPGTKVQHCHYCGGSGMETIN 265 Query: 255 TGPFLMXXXXXXXXXXXXIIKNPCHDCKGKGRVSQRKKIRVSIPAGIEDGQSLRVSLGAR 434 TGPF+M II NPC C+G G+ Q+K++ + +PAG+EDGQ++R+ +G R Sbjct: 266 TGPFVMRSTCRRCGGRGSIITNPCVVCRGAGQAKQKKRVTIPVPAGVEDGQTVRMPVGKR 325 Query: 435 SAQELYVTVRVEKSNLFRRDGSDVHSDITVSLPLAALGGKITVPGIYGDVTVDIPKGSCS 614 E++VT RV+KS +FRRDG+D+HSD+ +S+ A LGG G+Y + V IP G + Sbjct: 326 ---EIFVTFRVQKSPVFRRDGADIHSDLFISIAQAILGGTAKAQGLYETINVTIPAGIQT 382 Query: 615 HDRIRLPGKGIARVNSYGYGDHYLNIKIQPTKRLTETQRALLLALAEQD-ESAGMINGVT 791 +IRL GKGI R+NSYGYGDHY++IKI+ KRL+ Q+ L+L+ AE + + G +NGVT Sbjct: 383 DQKIRLTGKGIPRINSYGYGDHYIHIKIRVPKRLSSRQQNLILSYAEDETDVEGTVNGVT 442 Query: 792 MTANGKQAIDDT 827 T+ G + +D + Sbjct: 443 HTSTGGRTMDSS 454
>sp|Q24331|TID_DROVI Tumorous imaginal discs protein, mitochondrial precursor (Lethal(2)tumorous imaginal discs protein) (TID58) Length = 529 Score = 236 bits (603), Expect = 6e-62 Identities = 115/246 (46%), Positives = 174/246 (70%), Gaps = 1/246 (0%) Frame = +3 Query: 78 RAQEVNVNLTFGQAACGVNKELYVNLMMECGRCGGSKSEPGTSSTVCPSCQGSGVQSVNT 257 +AQE+ ++LTF QAA GVNK++ VN++ +C +C GSK EPGT C C G+G ++++T Sbjct: 214 QAQELVMDLTFAQAARGVNKDVNVNVVDQCPKCAGSKCEPGTKPGRCQYCNGTGFETIST 273 Query: 258 GPFLMXXXXXXXXXXXXIIKNPCHDCKGKGRVSQRKKIRVSIPAGIEDGQSLRVSLGARS 437 GPF+M IK PC +C+GKG+ QR+K+ V +PAGIE+GQ++R+ +G++ Sbjct: 274 GPFVMRSTCRYCQGTRQYIKYPCAECEGKGQTVQRRKVTVPVPAGIENGQTVRMQVGSK- 332 Query: 438 AQELYVTVRVEKSNLFRRDGSDVHSDITVSLPLAALGGKITVPGIYGDVTVDIPKGSCSH 617 EL+VT RVE+S+ FRRDG+DVH+D +SL A LGG + V G+Y D ++I G+ SH Sbjct: 333 --ELFVTFRVERSDYFRRDGADVHTDAPISLAQAVLGGTVRVQGVYEDQWLNIEPGTSSH 390 Query: 618 DRIRLPGKGIARVNSYGYGDHYLNIKIQPTKRLTETQRALLLALAE-QDESAGMINGVTM 794 +I L GKG+ RVN++G+GDHY++IKI+ K+L++ QRALL A AE ++++ G I+G+ Sbjct: 391 RKIALRGKGLKRVNAHGHGDHYVHIKIEVPKKLSQEQRALLEAYAELEEDTPGQIHGMAQ 450 Query: 795 TANGKQ 812 +G + Sbjct: 451 RKDGSK 456
>sp|Q27237|TID_DROME Tumorous imaginal discs protein, mitochondrial precursor (Lethal(2)tumorous imaginal discs protein) (TID56) (TID50) Length = 520 Score = 225 bits (573), Expect = 2e-58 Identities = 113/250 (45%), Positives = 173/250 (69%), Gaps = 4/250 (1%) Frame = +3 Query: 78 RAQEVNVNLTFGQAACGVNKELYVNLMMECGRCGGSKSEPGTSSTVCPSCQGSGVQSVNT 257 +AQE+ ++LTF QAA GVNK++ VN++ +C +C G+K EPGT C C G+G ++V+T Sbjct: 198 QAQEMVMDLTFAQAARGVNKDVNVNVVDQCPKCAGTKCEPGTKPGRCQYCNGTGFETVST 257 Query: 258 GPFLMXXXXXXXXXXXXIIKNPCHDCKGKGRVSQRKKIRVSIPAGIEDGQSLRVSLGARS 437 GPF+M IK PC +C+GKGR QR+K+ V +PAGIE+GQ++R+ +G++ Sbjct: 258 GPFVMRSTCRYCQGTRQHIKYPCSECEGKGRTVQRRKVTVPVPAGIENGQTVRMQVGSK- 316 Query: 438 AQELYVTVRVEKSNLFRRDGSDVHSDITVSLPLAALGGKITVPGIYGDVTVDIPKGSCSH 617 EL+VT RVE+S+ FRR+G+DVH+D +SL A LGG + V G+Y D +++ G+ SH Sbjct: 317 --ELFVTFRVERSDYFRREGADVHTDAAISLAQAVLGGTVRVQGVYEDQWINVEPGTSSH 374 Query: 618 DRIRLPGKGIARVNSYGYGDHYLNIKI--QPTKRLTETQRALLLALAE-QDESAGMINGV 788 +I L GKG+ RVN++G+GDHY+++KI K+L + + AL+ A AE ++++ G I+G+ Sbjct: 375 HKIMLRGKGLKRVNAHGHGDHYVHVKITVPSAKKLDKKRLALIEAYAELEEDTPGQIHGI 434 Query: 789 TMTANG-KQA 815 +G KQA Sbjct: 435 ANRKDGSKQA 444
>sp|Q8TA83|DNJ10_CAEEL DnaJ homolog dnj-10 (DnaJ domain protein 10) Length = 456 Score = 179 bits (453), Expect = 2e-44 Identities = 101/297 (34%), Positives = 160/297 (53%), Gaps = 3/297 (1%) Frame = +3 Query: 69 GPERAQEVNVNLTFGQAACGVNKELYVNLMMECGRCGGSKSEPGTSSTVCPSCQGSGVQS 248 G AQE+ ++++F +A G K + VN++ +C +C G++ EPG T CP C G+G S Sbjct: 159 GHSAAQEMVMDISFEEAVRGATKNVSVNVVEDCLKCHGTQVEPGHKKTSCPYCNGTGAVS 218 Query: 249 VNT-GPFLMXXXXXXXXXXXXIIKNPCHDCKGKGRVSQRKKIRVSIPAGIEDGQSLRVSL 425 G F KNPC +C+G+G+ QR+++ ++PAG +G SL+ + Sbjct: 219 QRLQGGFFYQTTCNRCRGSGHYNKNPCQECEGEGQTVQRRQVSFNVPAGTNNGDSLKFQV 278 Query: 426 GARSAQELYVTVRVEKSNLFRRDGSDVHSDITVSLPLAALGGKITVPGIYGDVTVDIPKG 605 G +L+V V S FRR+ D+H D+ +SL A LGG + VPGI GD V IP G Sbjct: 279 GKN---QLFVRFNVAPSLKFRREKDDIHCDVDISLAQAVLGGTVKVPGINGDTYVHIPAG 335 Query: 606 SCSHDRIRLPGKGIARVNSYGYGDHYLNIKIQPTKRLTETQRALLLALAEQDE-SAGMIN 782 + SH ++RL GKG+ R++SYG GD Y++IK+ K LT Q+ ++LA A ++ G I Sbjct: 336 TGSHTKMRLTGKGVKRLHSYGNGDQYMHIKVTVPKYLTAEQKQIMLAWAATEQLKDGTIK 395 Query: 783 GVTMTANGKQAIDDTDQFLLSQ-IRMVLKQHENTDANQPTNKQDQQQHENPSEKLKK 950 G+ ++ ++ S+ K+ A + Q++E EK+K+ Sbjct: 396 GLEKNQKTEEKETKKNEEKKSEGASESQKRRSEPVAENAETIDENQENEGFFEKIKR 452
>sp|O69269|DNAJ_BACSH Chaperone protein dnaJ Length = 368 Score = 161 bits (408), Expect = 3e-39 Identities = 87/233 (37%), Positives = 133/233 (57%), Gaps = 10/233 (4%) Frame = +3 Query: 87 EVNVNLTFGQAACGVNKELYVNLMMECGRCGGSKSEPGTSSTVCPSCQGSGV--QSVNT- 257 + +N+ F +A G E+ + C C GS ++PGT C +C G+G Q+V+T Sbjct: 112 QYRMNIKFEEAIFGKETEIEIPKDETCETCHGSGAKPGTQPETCSTCNGAGQINQAVDTP 171 Query: 258 -GPFLMXXXXXXXXXXXXIIKNPCHDCKGKGRVSQRKKIRVSIPAGIEDGQSLRVS---- 422 G + IIK C C+G+G+V +RKKI+VSIPAG++DGQ +RVS Sbjct: 172 FGRMMNRRSCTTCHGTGKIIKEKCSTCRGEGKVQKRKKIKVSIPAGVDDGQQIRVSGQGE 231 Query: 423 --LGARSAQELYVTVRVEKSNLFRRDGSDVHSDITVSLPLAALGGKITVPGIYGDVTVDI 596 + A +LY+ RV+ N F RDG D++ ++ ++ P AALG +I VP ++G V + I Sbjct: 232 PGINGGPAGDLYIMFRVQGHNDFERDGDDIYFELKLTFPQAALGDEIEVPTVHGKVKLRI 291 Query: 597 PKGSCSHDRIRLPGKGIARVNSYGYGDHYLNIKIQPTKRLTETQRALLLALAE 755 P G+ S + RL KG+ V+ YG G+ Y+ +K+ ++LTE Q+ LL AE Sbjct: 292 PAGTQSGAQFRLKDKGVKNVHGYGMGNQYVTVKVMTPEKLTEKQKQLLREFAE 344
>sp|Q892R1|DNAJ_CLOTE Chaperone protein dnaJ Length = 375 Score = 161 bits (407), Expect = 3e-39 Identities = 92/244 (37%), Positives = 133/244 (54%), Gaps = 12/244 (4%) Frame = +3 Query: 60 RDDGPERAQEV--NVNLTFGQAACGVNKELYVNLMMECGRCGGSKSEPGTSSTVCPSCQG 233 R +GP++ +V ++NLTF +A GV KE+ + C C G+ ++PGT+S CP C G Sbjct: 110 RRNGPQKGPDVQYSINLTFNEAVFGVEKEVSITKSETCETCTGTGAKPGTNSKTCPKCNG 169 Query: 234 SGVQSV--NT--GPFLMXXXXXXXXXXXXIIKNPCHDCKGKGRVSQRKKIRVSIPAGIED 401 SG V NT G F+ II +PC DCKGKG V ++KKI+V+IPAG++ Sbjct: 170 SGQIKVQRNTALGSFVSVNTCDMCGGKGTIISDPCSDCKGKGTVRKQKKIKVNIPAGVDT 229 Query: 402 GQSLRV------SLGARSAQELYVTVRVEKSNLFRRDGSDVHSDITVSLPLAALGGKITV 563 G + + + +LY++VRV F+R G D++ D +S A+LG ++ V Sbjct: 230 GNVIPIRGQGEAGVNGGPPGDLYISVRVMPDPFFKRRGDDIYIDQHISFAKASLGTELKV 289 Query: 564 PGIYGDVTVDIPKGSCSHDRIRLPGKGIARVNSYGYGDHYLNIKIQPTKRLTETQRALLL 743 I G+V DIP G+ RL GKG+ VN G GD Y+NI + K L + Q+ L Sbjct: 290 KTIDGEVKYDIPSGTQPGTVFRLKGKGVPHVNGRGRGDQYVNIIVDIPKSLNQKQKEALF 349 Query: 744 ALAE 755 A E Sbjct: 350 AYME 353
>sp|Q5KWZ8|DNAJ_GEOKA Chaperone protein dnaJ Length = 382 Score = 160 bits (405), Expect = 6e-39 Identities = 92/239 (38%), Positives = 128/239 (53%), Gaps = 12/239 (5%) Frame = +3 Query: 60 RDDGPERAQEVN--VNLTFGQAACGVNKELYVNLMMECGRCGGSKSEPGTSSTVCPSCQG 233 R GP + +V + LTF +AA G E+ + C C GS ++PGTS T CP C G Sbjct: 113 RASGPRKGADVEYMMTLTFEEAAFGKETEIEIpreETCDTCQGSGAKPGTSPTSCPHCHG 172 Query: 234 SG-VQSVNTGPF---LMXXXXXXXXXXXXIIKNPCHDCKGKGRVSQRKKIRVSIPAGIED 401 SG V S PF + I C C G GRV +RKKI V IPAG++D Sbjct: 173 SGQVTSEQATPFGRIVNRRTCPVCGGTGRYIPEKCPTCGGTGRVKRRKKIHVKIPAGVDD 232 Query: 402 GQSLRVS------LGARSAQELYVTVRVEKSNLFRRDGSDVHSDITVSLPLAALGGKITV 563 GQ LRV+ + +LY+ RVE F+RDG D++ ++ +S AALG +I V Sbjct: 233 GQQLRVAGQGEPGVNGGPPGDLYIIFRVEPHEFFKRDGDDIYCEVPLSFAQAALGDEIEV 292 Query: 564 PGIYGDVTVDIPKGSCSHDRIRLPGKGIARVNSYGYGDHYLNIKIQPTKRLTETQRALL 740 P ++G V + IP G+ + R RL GKG+ V YG GD ++ +++ +LTE Q+ LL Sbjct: 293 PTLHGHVKLKIPAGTQTGTRFRLKGKGVPNVRGYGQGDQHVIVRVVTPTKLTEKQKQLL 351
>sp|P63969|DNAJ_NEIMB Chaperone protein dnaJ sp|P63968|DNAJ_NEIMA Chaperone protein dnaJ Length = 373 Score = 159 bits (401), Expect = 2e-38 Identities = 83/224 (37%), Positives = 125/224 (55%), Gaps = 6/224 (2%) Frame = +3 Query: 87 EVNVNLTFGQAACGVNKELYVNLMMECGRCGGSKSEPGTSSTVCPSCQGSGVQSVNTGPF 266 +V + +T +AA GV K + + C C GS ++PGTS CP+C+GSG + F Sbjct: 121 QVGIEITLEEAAKGVKKRINIPTYEACDVCNGSGAKPGTSPETCPTCKGSGTVHIQQAIF 180 Query: 267 LMXXXXXXXXXXXXIIKNPCHDCKGKGRVSQRKKIRVSIPAGIEDGQSLRVS------LG 428 M IK PC C+G GR K + V+IPAGI+DGQ +R+S + Sbjct: 181 RMQQTCPTCHGAGKHIKEPCVKCRGAGRNKAVKTVEVNIPAGIDDGQRIRLSGEGGPGMH 240 Query: 429 ARSAQELYVTVRVEKSNLFRRDGSDVHSDITVSLPLAALGGKITVPGIYGDVTVDIPKGS 608 A +LYVTVR+ +F+RDG D+H ++ +S AALGG++ VP + G V + +PK + Sbjct: 241 GAPAGDLYVTVRIRAHKIFQRDGLDLHCELPISFATAALGGELEVPTLDGKVKLTVPKET 300 Query: 609 CSHDRIRLPGKGIARVNSYGYGDHYLNIKIQPTKRLTETQRALL 740 + R+R+ GKG+ + S GD Y +I ++ LT+ Q+ LL Sbjct: 301 QTGRRMRVKGKGVKSLRSSATGDLYCHIVVETPVNLTDRQKELL 344
>sp|Q5F5M1|DNAJ_NEIG1 Chaperone protein dnaJ Length = 373 Score = 157 bits (398), Expect = 4e-38 Identities = 82/224 (36%), Positives = 125/224 (55%), Gaps = 6/224 (2%) Frame = +3 Query: 87 EVNVNLTFGQAACGVNKELYVNLMMECGRCGGSKSEPGTSSTVCPSCQGSGVQSVNTGPF 266 +V + +T +AA GV K + + C C GS ++PG S CP+C+GSG + F Sbjct: 121 QVGIEITLEEAAKGVKKRINIPTYEACDVCNGSGAKPGASPETCPTCKGSGTVHIQQAIF 180 Query: 267 LMXXXXXXXXXXXXIIKNPCHDCKGKGRVSQRKKIRVSIPAGIEDGQSLRVS------LG 428 M IK PC C+G GR K + V+IPAGI+DGQ +R+S + Sbjct: 181 RMQQTCPTCRGAGKHIKEPCVKCRGVGRNKAVKTVEVNIPAGIDDGQRIRLSGEGGPGMH 240 Query: 429 ARSAQELYVTVRVEKSNLFRRDGSDVHSDITVSLPLAALGGKITVPGIYGDVTVDIPKGS 608 A +LYVTVR+ +F+RDG D+H ++ +S +AALGG++ VP + G V + +PK + Sbjct: 241 GAPAGDLYVTVRIRAHKIFQRDGLDLHCELPISFAMAALGGELEVPTLDGKVKLTVPKET 300 Query: 609 CSHDRIRLPGKGIARVNSYGYGDHYLNIKIQPTKRLTETQRALL 740 + R+R+ GKG+ + S GD Y +I ++ LT+ Q+ LL Sbjct: 301 QTGRRMRVKGKGVKSLRSSATGDLYCHIVVETPVNLTDRQKELL 344
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 117,251,550
Number of Sequences: 369166
Number of extensions: 2447494
Number of successful extensions: 7790
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6438
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6901
length of database: 68,354,980
effective HSP length: 111
effective length of database: 47,849,395
effective search space used: 11292457220
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail