DrC_00180
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
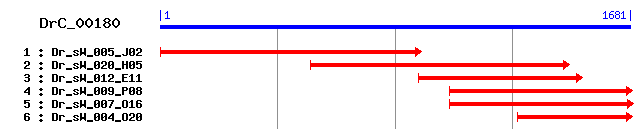
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00180 (1681 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O35286|DHX15_MOUSE Putative pre-mRNA splicing factor RNA... 916 0.0 sp|O43143|DHX15_HUMAN Putative pre-mRNA splicing factor RNA... 916 0.0 sp|Q20875|DHX15_CAEEL Putative pre-mRNA splicing factor ATP... 837 0.0 sp|O22899|DHX15_ARATH Putative pre-mRNA splicing factor ATP... 811 0.0 sp|O17438|DHX15_STRPU Putative pre-mRNA splicing factor ATP... 727 0.0 sp|P53131|PRP43_YEAST Pre-mRNA splicing factor RNA helicase... 686 0.0 sp|O42945|DHX15_SCHPO Probable pre-mRNA splicing factor RNA... 675 0.0 sp|Q38953|DHX8_ARATH Putative pre-mRNA splicing factor ATP-... 519 e-146 sp|Q14562|DHX8_HUMAN ATP-dependent helicase DHX8 (RNA helic... 518 e-146 sp|O42643|DHX8_SCHPO Putative pre-mRNA splicing factor ATP-... 506 e-143
>sp|O35286|DHX15_MOUSE Putative pre-mRNA splicing factor RNA helicase (DEAH box protein 15) Length = 795 Score = 916 bits (2367), Expect = 0.0 Identities = 441/532 (82%), Positives = 489/532 (91%) Frame = +3 Query: 39 LLDEAHERTLATDILMGLLKEVIKVRTDLKIVIMSATLDAGKFQEYFGNAPLMKVPGRTH 218 +LDEAHERTLATDILMG+LKEV++ R+DLK+++MSATLDAGKFQ YF N PL+ +PGRTH Sbjct: 258 ILDEAHERTLATDILMGVLKEVVRQRSDLKVIVMSATLDAGKFQIYFDNCPLLTIPGRTH 317 Query: 219 PVEIFYTPEPEKDYLEAAIRTVIQIHMCEESEGDILVFLTGQEEIDEACKRIQREVENLG 398 PVEIFYTPEPE+DYLEAAIRTVIQIHMCEE EGD+L+FLTGQEEIDEACKRI+REV++LG Sbjct: 318 PVEIFYTPEPERDYLEAAIRTVIQIHMCEEEEGDLLLFLTGQEEIDEACKRIKREVDDLG 377 Query: 399 PEVGELKCIPLYSTLPPNLQQRIFEPPPPKKANGAIGRKVVVSTNIAETSLTIDGVVFVI 578 PEVG++K IPLYSTLPP QQRIFEPPPPKK NGAIGRKVVVSTNIAETSLTIDGVVFVI Sbjct: 378 PEVGDIKIIPLYSTLPPQQQQRIFEPPPPKKQNGAIGRKVVVSTNIAETSLTIDGVVFVI 437 Query: 579 DPGFAKQKVFNPRIRVESLLVTPISKASXXXXXXXXXXTRPGKCFRLYTEKGYNNEMHEN 758 DPGFAKQKV+NPRIRVESLLVT ISKAS TRPGKCFRLYTEK Y EM +N Sbjct: 438 DPGFAKQKVYNPRIRVESLLVTAISKASAQQRAGRAGRTRPGKCFRLYTEKAYKTEMQDN 497 Query: 759 TYPEILRSNLGTVVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLAALDDDGNLTD 938 TYPEILRSNLG+VVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLAAL+DDG+LT+ Sbjct: 498 TYPEILRSNLGSVVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLAALNDDGDLTE 557 Query: 939 LGSMMAEFPLDPQLAKMVIASCDFNCSNEILSITAMLSVPQCFVRPTEARTAADEAKMGF 1118 LGSMMAEFPLDPQLAKMVIASCD+NCSNE+LSITAMLSVPQCFVRPTEA+ AADEAKM F Sbjct: 558 LGSMMAEFPLDPQLAKMVIASCDYNCSNEVLSITAMLSVPQCFVRPTEAKKAADEAKMRF 617 Query: 1119 AHIDGDHLTMLNVYHAFKQNREDSQWCYEHFVNYRSLKAADNVRIQLSRIMDRFNLRRSS 1298 AHIDGDHLT+LNVYHAFKQN E QWCY++F+NYRSL +ADNVR QLSRIMDRFNL R S Sbjct: 618 AHIDGDHLTLLNVYHAFKQNHESVQWCYDNFINYRSLMSADNVRQQLSRIMDRFNLPRRS 677 Query: 1299 TDFSSKDYYVNIRKALVSGFFMQVAHLERSGHYLTVKDNQIVHLHPSTCLDHKPEWVLYN 1478 TDF+S+DYY+NIRKALV+G+FMQVAHLER+GHYLTVKDNQ+V LHPST LDHKPEWVLYN Sbjct: 678 TDFTSRDYYINIRKALVTGYFMQVAHLERTGHYLTVKDNQVVQLHPSTVLDHKPEWVLYN 737 Query: 1479 EFVLTTKNFIRTVCEVKPEWLVKVAPQYYDMSNFPPCAAKEIIERIINRMQN 1634 EFVLTTKN+IRT ++KPEWLVK+APQYYDMSNFP C AK ++RII ++Q+ Sbjct: 738 EFVLTTKNYIRTCTDIKPEWLVKIAPQYYDMSNFPQCEAKRQLDRIIAKLQS 789
>sp|O43143|DHX15_HUMAN Putative pre-mRNA splicing factor RNA helicase (DEAH box protein 15) (ATP-dependent RNA helicase #46) Length = 795 Score = 916 bits (2367), Expect = 0.0 Identities = 441/532 (82%), Positives = 489/532 (91%) Frame = +3 Query: 39 LLDEAHERTLATDILMGLLKEVIKVRTDLKIVIMSATLDAGKFQEYFGNAPLMKVPGRTH 218 +LDEAHERTLATDILMG+LKEV++ R+DLK+++MSATLDAGKFQ YF N PL+ +PGRTH Sbjct: 258 ILDEAHERTLATDILMGVLKEVVRQRSDLKVIVMSATLDAGKFQIYFDNCPLLTIPGRTH 317 Query: 219 PVEIFYTPEPEKDYLEAAIRTVIQIHMCEESEGDILVFLTGQEEIDEACKRIQREVENLG 398 PVEIFYTPEPE+DYLEAAIRTVIQIHMCEE EGD+L+FLTGQEEIDEACKRI+REV++LG Sbjct: 318 PVEIFYTPEPERDYLEAAIRTVIQIHMCEEEEGDLLLFLTGQEEIDEACKRIKREVDDLG 377 Query: 399 PEVGELKCIPLYSTLPPNLQQRIFEPPPPKKANGAIGRKVVVSTNIAETSLTIDGVVFVI 578 PEVG++K IPLYSTLPP QQRIFEPPPPKK NGAIGRKVVVSTNIAETSLTIDGVVFVI Sbjct: 378 PEVGDIKIIPLYSTLPPQQQQRIFEPPPPKKQNGAIGRKVVVSTNIAETSLTIDGVVFVI 437 Query: 579 DPGFAKQKVFNPRIRVESLLVTPISKASXXXXXXXXXXTRPGKCFRLYTEKGYNNEMHEN 758 DPGFAKQKV+NPRIRVESLLVT ISKAS TRPGKCFRLYTEK Y EM +N Sbjct: 438 DPGFAKQKVYNPRIRVESLLVTAISKASAQQRAGRAGRTRPGKCFRLYTEKAYKTEMQDN 497 Query: 759 TYPEILRSNLGTVVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLAALDDDGNLTD 938 TYPEILRSNLG+VVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLAAL+DDG+LT+ Sbjct: 498 TYPEILRSNLGSVVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLAALNDDGDLTE 557 Query: 939 LGSMMAEFPLDPQLAKMVIASCDFNCSNEILSITAMLSVPQCFVRPTEARTAADEAKMGF 1118 LGSMMAEFPLDPQLAKMVIASCD+NCSNE+LSITAMLSVPQCFVRPTEA+ AADEAKM F Sbjct: 558 LGSMMAEFPLDPQLAKMVIASCDYNCSNEVLSITAMLSVPQCFVRPTEAKKAADEAKMRF 617 Query: 1119 AHIDGDHLTMLNVYHAFKQNREDSQWCYEHFVNYRSLKAADNVRIQLSRIMDRFNLRRSS 1298 AHIDGDHLT+LNVYHAFKQN E QWCY++F+NYRSL +ADNVR QLSRIMDRFNL R S Sbjct: 618 AHIDGDHLTLLNVYHAFKQNHESVQWCYDNFINYRSLMSADNVRQQLSRIMDRFNLPRRS 677 Query: 1299 TDFSSKDYYVNIRKALVSGFFMQVAHLERSGHYLTVKDNQIVHLHPSTCLDHKPEWVLYN 1478 TDF+S+DYY+NIRKALV+G+FMQVAHLER+GHYLTVKDNQ+V LHPST LDHKPEWVLYN Sbjct: 678 TDFTSRDYYINIRKALVTGYFMQVAHLERTGHYLTVKDNQVVQLHPSTVLDHKPEWVLYN 737 Query: 1479 EFVLTTKNFIRTVCEVKPEWLVKVAPQYYDMSNFPPCAAKEIIERIINRMQN 1634 EFVLTTKN+IRT ++KPEWLVK+APQYYDMSNFP C AK ++RII ++Q+ Sbjct: 738 EFVLTTKNYIRTCTDIKPEWLVKIAPQYYDMSNFPQCEAKRQLDRIIAKLQS 789
>sp|Q20875|DHX15_CAEEL Putative pre-mRNA splicing factor ATP-dependent RNA helicase F56D2.6 Length = 739 Score = 837 bits (2163), Expect = 0.0 Identities = 402/531 (75%), Positives = 460/531 (86%) Frame = +3 Query: 39 LLDEAHERTLATDILMGLLKEVIKVRTDLKIVIMSATLDAGKFQEYFGNAPLMKVPGRTH 218 +LDEAHERTLATDILMGL+KE+++ R D+K+VIMSATLDAGKFQ YF + PL+ VPGRT Sbjct: 202 ILDEAHERTLATDILMGLIKEIVRNRADIKVVIMSATLDAGKFQRYFEDCPLLSVPGRTF 261 Query: 219 PVEIFYTPEPEKDYLEAAIRTVIQIHMCEESEGDILVFLTGQEEIDEACKRIQREVENLG 398 PVEIF+TP EKDYLEAAIRTVIQIHM EE EGDIL+FLTGQEEI+EACKRI RE++ LG Sbjct: 262 PVEIFFTPNAEKDYLEAAIRTVIQIHMVEEVEGDILLFLTGQEEIEEACKRIDREIQALG 321 Query: 399 PEVGELKCIPLYSTLPPNLQQRIFEPPPPKKANGAIGRKVVVSTNIAETSLTIDGVVFVI 578 + G L CIPLYSTLPP QQRIFEP PP + NGAI RK V+STNIAETSLTIDGVVFVI Sbjct: 322 ADAGALSCIPLYSTLPPAAQQRIFEPAPPNRPNGAISRKCVISTNIAETSLTIDGVVFVI 381 Query: 579 DPGFAKQKVFNPRIRVESLLVTPISKASXXXXXXXXXXTRPGKCFRLYTEKGYNNEMHEN 758 DPGF+KQKV+NPRIRVESLLV PISKAS T+PGKCFRLYTE Y +EM + Sbjct: 382 DPGFSKQKVYNPRIRVESLLVCPISKASAMQRAGRAGRTKPGKCFRLYTETAYGSEMQDQ 441 Query: 759 TYPEILRSNLGTVVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLAALDDDGNLTD 938 TYPEILRSNLG+VVLQLKKLG +DLVHFDFMDPPAPETLMRALELLNYL A++DDG LT+ Sbjct: 442 TYPEILRSNLGSVVLQLKKLGTEDLVHFDFMDPPAPETLMRALELLNYLQAINDDGELTE 501 Query: 939 LGSMMAEFPLDPQLAKMVIASCDFNCSNEILSITAMLSVPQCFVRPTEARTAADEAKMGF 1118 LGS+MAEFPLDPQLAKM+I S + NCSNEILSITAMLSVPQC+VRP E RT ADEAK F Sbjct: 502 LGSLMAEFPLDPQLAKMLITSTELNCSNEILSITAMLSVPQCWVRPNEMRTEADEAKARF 561 Query: 1119 AHIDGDHLTMLNVYHAFKQNREDSQWCYEHFVNYRSLKAADNVRIQLSRIMDRFNLRRSS 1298 AHIDGDHLT+LNVYH+FKQN+ED QWCY++F+NYR++K AD VR QLSR+MD++NLRR S Sbjct: 562 AHIDGDHLTLLNVYHSFKQNQEDPQWCYDNFINYRTMKTADTVRTQLSRVMDKYNLRRVS 621 Query: 1299 TDFSSKDYYVNIRKALVSGFFMQVAHLERSGHYLTVKDNQIVHLHPSTCLDHKPEWVLYN 1478 TDF S+DYY+NIRKALV+GFFMQVAHLERSGHY+TVKDNQ+V+LHPST LDHKPEW LYN Sbjct: 622 TDFKSRDYYLNIRKALVAGFFMQVAHLERSGHYVTVKDNQLVNLHPSTVLDHKPEWALYN 681 Query: 1479 EFVLTTKNFIRTVCEVKPEWLVKVAPQYYDMSNFPPCAAKEIIERIINRMQ 1631 EFVLTTKNFIRTV +V+PEWL+++APQYYD+ NFP K + ++ +Q Sbjct: 682 EFVLTTKNFIRTVTDVRPEWLLQIAPQYYDLDNFPDGDTKRKLTTVMQTLQ 732
>sp|O22899|DHX15_ARATH Putative pre-mRNA splicing factor ATP-dependent RNA helicase Length = 729 Score = 811 bits (2094), Expect = 0.0 Identities = 388/535 (72%), Positives = 456/535 (85%), Gaps = 1/535 (0%) Frame = +3 Query: 39 LLDEAHERTLATDILMGLLKEVIKVRTDLKIVIMSATLDAGKFQEYFGNAPLMKVPGRTH 218 +LDEAHERTLATD+L GLLKEV++ R DLK+V+MSATL+A KFQEYF APLMKVPGR H Sbjct: 189 ILDEAHERTLATDVLFGLLKEVLRNRPDLKLVVMSATLEAEKFQEYFSGAPLMKVPGRLH 248 Query: 219 PVEIFYTPEPEKDYLEAAIRTVIQIHMCEESEGDILVFLTGQEEIDEACKRIQREVENLG 398 PVEIFYT EPE+DYLEAAIRTV+QIHMCE GDILVFLTG+EEI++AC++I +EV NLG Sbjct: 249 PVEIFYTQEPERDYLEAAIRTVVQIHMCEPP-GDILVFLTGEEEIEDACRKINKEVSNLG 307 Query: 399 PEVGELKCIPLYSTLPPNLQQRIFEPPP-PKKANGAIGRKVVVSTNIAETSLTIDGVVFV 575 +VG +K +PLYSTLPP +QQ+IF+P P P G GRK+VVSTNIAETSLTIDG+V+V Sbjct: 308 DQVGPVKVVPLYSTLPPAMQQKIFDPAPVPLTEGGPAGRKIVVSTNIAETSLTIDGIVYV 367 Query: 576 IDPGFAKQKVFNPRIRVESLLVTPISKASXXXXXXXXXXTRPGKCFRLYTEKGYNNEMHE 755 IDPGFAKQKV+NPRIRVESLLV+PISKAS TRPGKCFRLYTEK +NN++ Sbjct: 368 IDPGFAKQKVYNPRIRVESLLVSPISKASAHQRSGRAGRTRPGKCFRLYTEKSFNNDLQP 427 Query: 756 NTYPEILRSNLGTVVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLAALDDDGNLT 935 TYPEILRSNL VL LKKLGIDDLVHFDFMDPPAPETLMRALE+LNYL ALDD+GNLT Sbjct: 428 QTYPEILRSNLANTVLTLKKLGIDDLVHFDFMDPPAPETLMRALEVLNYLGALDDEGNLT 487 Query: 936 DLGSMMAEFPLDPQLAKMVIASCDFNCSNEILSITAMLSVPQCFVRPTEARTAADEAKMG 1115 G +M+EFPLDPQ++KM+I S +FNCSNEILS++AMLSVP CFVRP EA+ AADEAK Sbjct: 488 KTGEIMSEFPLDPQMSKMLIVSPEFNCSNEILSVSAMLSVPNCFVRpreAQKAADEAKAR 547 Query: 1116 FAHIDGDHLTMLNVYHAFKQNREDSQWCYEHFVNYRSLKAADNVRIQLSRIMDRFNLRRS 1295 F HIDGDHLT+LNVYHA+KQN ED WC+E+FVN R++K+ADNVR QL RIM RFNL+ Sbjct: 548 FGHIDGDHLTLLNVYHAYKQNNEDPNWCFENFVNNRAMKSADNVRQQLVRIMSRFNLKMC 607 Query: 1296 STDFSSKDYYVNIRKALVSGFFMQVAHLERSGHYLTVKDNQIVHLHPSTCLDHKPEWVLY 1475 STDF+S+DYYVNIRKA+++G+FMQVAHLER+GHYLTVKDNQ+VHLHPS CLDHKPEWV+Y Sbjct: 608 STDFNSRDYYVNIRKAMLAGYFMQVAHLERTGHYLTVKDNQVVHLHPSNCLDHKPEWVIY 667 Query: 1476 NEFVLTTKNFIRTVCEVKPEWLVKVAPQYYDMSNFPPCAAKEIIERIINRMQNGK 1640 NE+VLTT+NFIRTV +++ EWLV VA YYD+SNFP C AK +E++ + + K Sbjct: 668 NEYVLTTRNFIRTVTDIRGEWLVDVAQHYYDLSNFPNCEAKRALEKLYKKREREK 722
>sp|O17438|DHX15_STRPU Putative pre-mRNA splicing factor ATP-dependent RNA helicase PRP1 Length = 455 Score = 727 bits (1876), Expect = 0.0 Identities = 358/430 (83%), Positives = 394/430 (91%) Frame = +3 Query: 39 LLDEAHERTLATDILMGLLKEVIKVRTDLKIVIMSATLDAGKFQEYFGNAPLMKVPGRTH 218 LLDEAHERT+ATDILMGLLKEV K R+DLK+V+MSATLDAGKFQ YF NAPLM VPGRTH Sbjct: 26 LLDEAHERTVATDILMGLLKEVEKQRSDLKLVVMSATLDAGKFQHYFDNAPLMTVPGRTH 85 Query: 219 PVEIFYTPEPEKDYLEAAIRTVIQIHMCEESEGDILVFLTGQEEIDEACKRIQREVENLG 398 PVEIFYTPEPE+DYLEAAIRTV+QIHMCEE EGD+L+FLTGQEEI+EACKRI+REV+NLG Sbjct: 86 PVEIFYTPEPERDYLEAAIRTVVQIHMCEEVEGDVLLFLTGQEEIEEACKRIKREVDNLG 145 Query: 399 PEVGELKCIPLYSTLPPNLQQRIFEPPPPKKANGAIGRKVVVSTNIAETSLTIDGVVFVI 578 PEVG+LK IPLYSTLPP +QQRIFE PP KANGAIGRKVVVSTNIAETSLTIDGVVFVI Sbjct: 146 PEVGDLKTIPLYSTLPPAMQQRIFEHAPPNKANGAIGRKVVVSTNIAETSLTIDGVVFVI 205 Query: 579 DPGFAKQKVFNPRIRVESLLVTPISKASXXXXXXXXXXTRPGKCFRLYTEKGYNNEMHEN 758 DPGFAKQKV+NPRIRVESLLV+PISKAS TRPGKCFRLYTEK Y++EM +N Sbjct: 206 DPGFAKQKVYNPRIRVESLLVSPISKASAQQRVGRAGRTRPGKCFRLYTEKAYDSEMQDN 265 Query: 759 TYPEILRSNLGTVVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLAALDDDGNLTD 938 TYPEILRSNLGTVVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYL ALDD G+LT Sbjct: 266 TYPEILRSNLGTVVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLGALDDSGDLTR 325 Query: 939 LGSMMAEFPLDPQLAKMVIASCDFNCSNEILSITAMLSVPQCFVRPTEARTAADEAKMGF 1118 LGSMMAEFPLDPQLAKMVIAS D++CSNEILS+TAMLSVPQCF+RP EA+ ADEAKM F Sbjct: 326 LGSMMAEFPLDPQLAKMVIASTDYSCSNEILSVTAMLSVPQCFLRPNEAKKLADEAKMRF 385 Query: 1119 AHIDGDHLTMLNVYHAFKQNREDSQWCYEHFVNYRSLKAADNVRIQLSRIMDRFNLRRSS 1298 AHIDGDHLT+LNVYHAFKQN ED QWCY++F+ YRSLK+AD+VR QL+RIMDRF L+R+S Sbjct: 386 AHIDGDHLTLLNVYHAFKQNNEDPQWCYDNFIQYRSLKSADSVRQQLARIMDRFALQRTS 445 Query: 1299 TDFSSKDYYV 1328 T+F+SKDYY+ Sbjct: 446 TNFNSKDYYL 455
>sp|P53131|PRP43_YEAST Pre-mRNA splicing factor RNA helicase PRP43 (Helicase JA1) Length = 767 Score = 686 bits (1769), Expect = 0.0 Identities = 349/548 (63%), Positives = 430/548 (78%), Gaps = 14/548 (2%) Frame = +3 Query: 39 LLDEAHERTLATDILMGLLKEVIKVRTDLKIVIMSATLDAGKFQEYFGNAPLMKVPGRTH 218 +LDEAHERTLATDILMGLLK+V+K R DLKI+IMSATLDA KFQ YF +APL+ VPGRT+ Sbjct: 213 ILDEAHERTLATDILMGLLKQVVKRRPDLKIIIMSATLDAEKFQRYFNDAPLLAVPGRTY 272 Query: 219 PVEIFYTPEPEKDYLEAAIRTVIQIHMCEESEGDILVFLTGQEEIDEACKRIQREVENLG 398 PVE++YTPE ++DYL++AIRTV+QIH EE+ GDIL+FLTG++EI++A ++I E + L Sbjct: 273 PVELYYTPEFQRDYLDSAIRTVLQIHATEEA-GDILLFLTGEDEIEDAVRKISLEGDQLV 331 Query: 399 PE--VGELKCIPLYSTLPPNLQQRIFEPPPPKKANGAIGRKVVVSTNIAETSLTIDGVVF 572 E G L PLY +LPP+ QQRIFEP P + NG GRKVV+STNIAETSLTIDG+V+ Sbjct: 332 REEGCGPLSVYPLYGSLPPHQQQRIFEPAP-ESHNGRPGRKVVISTNIAETSLTIDGIVY 390 Query: 573 VIDPGFAKQKVFNPRIRVESLLVTPISKASXXXXXXXXXXTRPGKCFRLYTEKGYNNEMH 752 V+DPGF+KQKV+NPRIRVESLLV+PISKAS TRPGKCFRLYTE+ + E+ Sbjct: 391 VVDPGFSKQKVYNPRIRVESLLVSPISKASAQQRAGRAGRTRPGKCFRLYTEEAFQKELI 450 Query: 753 ENTYPEILRSNLGTVVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLAALDDDGNL 932 E +YPEILRSNL + VL+LKKLGIDDLVHFDFMDPPAPET+MRALE LNYLA LDD+GNL Sbjct: 451 EQSYPEILRSNLSSTVLELKKLGIDDLVHFDFMDPPAPETMMRALEELNYLACLDDEGNL 510 Query: 933 TDLGSMMAEFPLDPQLAKMVIASCDFNCSNEILSITAMLSVPQCFVRPTEARTAADEAKM 1112 T LG + ++FPLDP LA M+I S +F CS EIL+I AMLSVP F+RPT+ + AD+AK Sbjct: 511 TPLGRLASQFPLDPMLAVMLIGSFEFQCSQEILTIVAMLSVPNVFIRPTKDKKRADDAKN 570 Query: 1113 GFAHIDGDHLTMLNVYHAFKQNREDS----QWCYEHFVNYRSLKAADNVRIQLSRIMDRF 1280 FAH DGDH+T+LNVYHAFK + +WC +H++NYRSL AADN+R QL R+M+R+ Sbjct: 571 IFAHPDGDHITLLNVYHAFKSDEAYEYGIHKWCRDHYLNYRSLSAADNIRSQLERLMNRY 630 Query: 1281 NLRRSSTDFSSKDYYVNIRKALVSGFFMQVAHLERSG--HYLTVKDNQIVHLHPSTCLDH 1454 NL ++TD+ S Y+ NIRKAL SGFFMQVA +RSG Y+TVKDNQ V +HPST L H Sbjct: 631 NLELNTTDYESPKYFDNIRKALASGFFMQVAK-KRSGAKGYITVKDNQDVLIHPSTVLGH 689 Query: 1455 KPEWVLYNEFVLTTKNFIRTVCEVKPEWLVKVAPQYYDMSNFPPCAAKEIIERI------ 1616 EWV+YNEFVLT+KN+IRTV V+PEWL+++AP YYD+SNF K +ERI Sbjct: 690 DAEWVIYNEFVLTSKNYIRTVTSVRPEWLIEIAPAYYDLSNFQKGDVKLSLERIKEKVDR 749 Query: 1617 INRMQNGK 1640 +N ++ GK Sbjct: 750 LNELKQGK 757
>sp|O42945|DHX15_SCHPO Probable pre-mRNA splicing factor RNA helicase prp43 Length = 735 Score = 675 bits (1742), Expect = 0.0 Identities = 336/517 (64%), Positives = 410/517 (79%), Gaps = 3/517 (0%) Frame = +3 Query: 39 LLDEAHERTLATDILMGLLKEVIKVRTDLKIVIMSATLDAGKFQEYFGNAPLMKVPGRTH 218 +LDEAHERTLATDILMGL+K + R DLKI++MSATLDA KFQ+YF +APL+ VPGRT+ Sbjct: 195 ILDEAHERTLATDILMGLMKRLATRRPDLKIIVMSATLDAKKFQKYFFDAPLLAVPGRTY 254 Query: 219 PVEIFYTPEPEKDYLEAAIRTVIQIHMCEESEGDILVFLTGQEEIDEACKRIQREVENLG 398 PVEI+YT EPE+DYLEAA+RTV+QIH+ EE GDILVFLTG+EEI++AC++I E ++L Sbjct: 255 PVEIYYTQEPERDYLEAALRTVLQIHV-EEGPGDILVFLTGEEEIEDACRKITLEADDLV 313 Query: 399 PE--VGELKCIPLYSTLPPNLQQRIFEPPPPKKANGAIGRKVVVSTNIAETSLTIDGVVF 572 E G LK PLY +LPPN QQRIFEP P +G GRKVV+STNIAETSLTIDG+V+ Sbjct: 314 REGAAGPLKVYPLYGSLPPNQQQRIFEPTPEDTKSG-YGRKVVISTNIAETSLTIDGIVY 372 Query: 573 VIDPGFAKQKVFNPRIRVESLLVTPISKASXXXXXXXXXXTRPGKCFRLYTEKGYNNEMH 752 V+DPGF+KQK++NPRIRVESLLV+PISKAS TRPGKCFRLYTE+ + E+ Sbjct: 373 VVDPGFSKQKIYNPRIRVESLLVSPISKASAQQRAGRAGRTRPGKCFRLYTEEAFRKELI 432 Query: 753 ENTYPEILRSNLGTVVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLAALDDDGNL 932 E TYPEILRSNL + VL+LKKLGIDDLVHFD+MDPPAPET+MRALE LNYL LDD+G+L Sbjct: 433 EQTYPEILRSNLSSTVLELKKLGIDDLVHFDYMDPPAPETMMRALEELNYLNCLDDNGDL 492 Query: 933 TDLGSMMAEFPLDPQLAKMVIASCDFNCSNEILSITAMLSVPQCFVRPTEARTAADEAKM 1112 T LG +EFPLDP LA M+I S +F CSNE+LS+TA+LSVP FVRP AR ADE + Sbjct: 493 TPLGRKASEFPLDPNLAVMLIRSPEFYCSNEVLSLTALLSVPNVFVRPNSARKLADEMRQ 552 Query: 1113 GFAHIDGDHLTMLNVYHAFKQNREDSQWCYEHFVNYRSLKAADNVRIQLSRIMDRFNLRR 1292 F H DGDHLT+LNVYHA+K + WC+ HF+++R+L +ADNVR QL R M+R + Sbjct: 553 QFTHPDGDHLTLLNVYHAYKSGEGTADWCWNHFLSHRALISADNVRKQLRRTMERQEVEL 612 Query: 1293 SSTDFSSKDYYVNIRKALVSGFFMQVAHLERSG-HYLTVKDNQIVHLHPSTCLDHKPEWV 1469 ST F K+YYVNIR+ALVSGFFMQVA +G +Y+T+KDNQ+V LHPS L PEWV Sbjct: 613 ISTPFDDKNYYVNIRRALVSGFFMQVAKKSANGKNYVTMKDNQVVSLHPSCGLSVTPEWV 672 Query: 1470 LYNEFVLTTKNFIRTVCEVKPEWLVKVAPQYYDMSNF 1580 +YNEFVLTTK+FIR V ++PEWL+++AP YYD+ +F Sbjct: 673 VYNEFVLTTKSFIRNVTAIRPEWLIELAPNYYDLDDF 709
>sp|Q38953|DHX8_ARATH Putative pre-mRNA splicing factor ATP-dependent RNA helicase Length = 1168 Score = 519 bits (1336), Expect = e-146 Identities = 263/527 (49%), Positives = 361/527 (68%), Gaps = 1/527 (0%) Frame = +3 Query: 39 LLDEAHERTLATDILMGLLKEVIKVRTDLKIVIMSATLDAGKFQEYFGNAPLMKVPGRTH 218 +LDEAHERT+ TD+L GLLK+++K R DL++++ SATLDA KF YF N + +PGRT Sbjct: 633 MLDEAHERTIHTDVLFGLLKKLMKRRLDLRLIVTSATLDAEKFSGYFFNCNIFTIPGRTF 692 Query: 219 PVEIFYTPEPEKDYLEAAIRTVIQIHMCEESEGDILVFLTGQEEIDEACKRIQREVENLG 398 PVEI YT +PE DYL+AA+ TV+QIH+ E EGDILVFLTGQEEID AC+ + ++ LG Sbjct: 693 PVEILYTKQPETDYLDAALITVLQIHLTEP-EGDILVFLTGQEEIDSACQSLYERMKGLG 751 Query: 399 PEVGELKCIPLYSTLPPNLQQRIFEPPPPKKANGAIGRKVVVSTNIAETSLTIDGVVFVI 578 V EL +P+YS LP +Q RIF+PPPP K RKVVV+TNIAE SLTIDG+ +V+ Sbjct: 752 KNVPELIILPVYSALPSEMQSRIFDPPPPGK------RKVVVATNIAEASLTIDGIYYVV 805 Query: 579 DPGFAKQKVFNPRIRVESLLVTPISKASXXXXXXXXXXTRPGKCFRLYTEKGYNNEMHEN 758 DPGFAKQ V+NP+ +ESL++TPIS+AS T PGKC+RLYTE Y NEM Sbjct: 806 DPGFAKQNVYNPKQGLESLVITPISQASAKQRAGRAGRTGPGKCYRLYTESAYRNEMPPT 865 Query: 759 TYPEILRSNLGTVVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLAALDDDGNLTD 938 + PEI R NLG L +K +GI+DL+ FDFMDPP P+ L+ A+E L L ALD++G LT Sbjct: 866 SIPEIQRINLGMTTLTMKAMGINDLLSFDFMDPPQPQALISAMEQLYSLGALDEEGLLTK 925 Query: 939 LGSMMAEFPLDPQLAKMVIASCDFNCSNEILSITAMLSVPQCFVRPTEARTAADEAKMGF 1118 LG MAEFPL+P L+KM++AS D CS+EIL++ AM+ F RP E + AD+ + F Sbjct: 926 LGRKMAEFPLEPPLSKMLLASVDLGCSDEILTMIAMIQTGNIFYRpreKQAQADQKRAKF 985 Query: 1119 AHIDGDHLTMLNVYHAFKQNREDSQWCYEHFVNYRSLKAADNVRIQLSRIMDRFNLRRSS 1298 +GDHLT+L VY A+K WC+E+F+ RSL+ A +VR QL IMD++ L Sbjct: 986 FQPEGDHLTLLAVYEAWKAKNFSGPWCFENFIQSRSLRRAQDVRKQLLSIMDKYKL---- 1041 Query: 1299 TDFSSKDYYVNIRKALVSGFFMQVAHLERSGHYLTVKDNQIVHLHPSTCL-DHKPEWVLY 1475 ++ + IRKA+ +GFF A + Y T+ +NQ V++HPS+ L +P+WV+Y Sbjct: 1042 DVVTAGKNFTKIRKAITAGFFFHGARKDPQEGYRTLVENQPVYIHPSSALFQRQPDWVIY 1101 Query: 1476 NEFVLTTKNFIRTVCEVKPEWLVKVAPQYYDMSNFPPCAAKEIIERI 1616 ++ V+TTK ++R V + P+WLV++AP+++ +S+ + ++ ERI Sbjct: 1102 HDLVMTTKEYMREVTVIDPKWLVELAPRFFKVSDPTKMSKRKRQERI 1148
>sp|Q14562|DHX8_HUMAN ATP-dependent helicase DHX8 (RNA helicase HRH1) (DEAH-box protein 8) Length = 1220 Score = 518 bits (1335), Expect = e-146 Identities = 262/514 (50%), Positives = 356/514 (69%), Gaps = 1/514 (0%) Frame = +3 Query: 39 LLDEAHERTLATDILMGLLKEVIKVRTDLKIVIMSATLDAGKFQEYFGNAPLMKVPGRTH 218 +LDEAHERT+ TD+L GLLK+ ++ R D+K+++ SATLDA KF +YF AP+ +PGRT+ Sbjct: 683 MLDEAHERTIHTDVLFGLLKKTVQKRQDMKLIVTSATLDAVKFSQYFYEAPIFTIPGRTY 742 Query: 219 PVEIFYTPEPEKDYLEAAIRTVIQIHMCEESEGDILVFLTGQEEIDEACKRIQREVENLG 398 PVEI YT EPE DYL+A++ TV+QIH+ E GDILVFLTGQEEID AC+ + +++LG Sbjct: 743 PVEILYTKEPETDYLDASLITVMQIHLTEPP-GDILVFLTGQEEIDTACEILYERMKSLG 801 Query: 399 PEVGELKCIPLYSTLPPNLQQRIFEPPPPKKANGAIGRKVVVSTNIAETSLTIDGVVFVI 578 P+V EL +P+YS LP +Q RIF+P PP RKVV++TNIAETSLTIDG+ +V+ Sbjct: 802 PDVPELIILPVYSALPSEMQTRIFDPAPPGS------RKVVIATNIAETSLTIDGIYYVV 855 Query: 579 DPGFAKQKVFNPRIRVESLLVTPISKASXXXXXXXXXXTRPGKCFRLYTEKGYNNEMHEN 758 DPGF KQKV+N + ++ L+VTPIS+A T PGKC+RLYTE+ Y +EM Sbjct: 856 DPGFVKQKVYNSKTGIDQLVVTPISQAQAKQRAGRAGRTGPGKCYRLYTERAYRDEMLTT 915 Query: 759 TYPEILRSNLGTVVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLAALDDDGNLTD 938 PEI R+NL + VL LK +GI+DL+ FDFMD P ETL+ A+E L L ALDD+G LT Sbjct: 916 NVPEIQRTNLASTVLSLKAMGINDLLSFDFMDAPPMETLITAMEQLYTLGALDDEGLLTR 975 Query: 939 LGSMMAEFPLDPQLAKMVIASCDFNCSNEILSITAMLSVPQCFVRPTEARTAADEAKMGF 1118 LG MAEFPL+P L KM+I S CS E+L+I +MLSV F RP + + AD+ K F Sbjct: 976 LGRRMAEFPLEPMLCKMLIMSVHLGCSEEMLTIVSMLSVQNVFYRPKDKQALADQKKAKF 1035 Query: 1119 AHIDGDHLTMLNVYHAFKQNREDSQWCYEHFVNYRSLKAADNVRIQLSRIMDRFNLRRSS 1298 +GDHLT+L VY+++K N+ + WCYE+F+ RSL+ A ++R Q+ IMDR L S Sbjct: 1036 HQTEGDHLTLLAVYNSWKNNKFSNPWCYENFIQARSLRRAQDIRKQMLGIMDRHKLDVVS 1095 Query: 1299 TDFSSKDYYVNIRKALVSGFFMQVAHLERSGHYLTVKDNQIVHLHPSTCL-DHKPEWVLY 1475 S+ V ++KA+ SGFF A + Y T+ D Q+V++HPS+ L + +PEWV+Y Sbjct: 1096 CGKST----VRVQKAICSGFFRNAAKKDPQEGYRTLIDQQVVYIHPSSALFNRQPEWVVY 1151 Query: 1476 NEFVLTTKNFIRTVCEVKPEWLVKVAPQYYDMSN 1577 +E VLTTK ++R V + P WLV+ AP ++ +S+ Sbjct: 1152 HELVLTTKEYMREVTTIDPRWLVEFAPAFFKVSD 1185
>sp|O42643|DHX8_SCHPO Putative pre-mRNA splicing factor ATP-dependent RNA helicase C10F6.02c Length = 1168 Score = 506 bits (1304), Expect = e-143 Identities = 259/515 (50%), Positives = 350/515 (67%), Gaps = 2/515 (0%) Frame = +3 Query: 39 LLDEAHERTLATDILMGLLKEVIKVRTDLKIVIMSATLDAGKFQEYFGNAPLMKVPGRTH 218 +LDEAHERT+ATD+L GLLK + R DLK+++ SATLDA +F YF P+ +PGR++ Sbjct: 629 ILDEAHERTVATDVLFGLLKGTVLKRPDLKLIVTSATLDAERFSSYFYKCPIFTIPGRSY 688 Query: 219 PVEIFYTPEPEKDYLEAAIRTVIQIHMCEESEGDILVFLTGQEEIDEACKRIQREVENLG 398 PVEI YT +PE DYL+AA+ TV+QIH+ E GDILVFLTGQEEID +C+ + + LG Sbjct: 689 PVEIMYTKQPEADYLDAALMTVMQIHL-SEGPGDILVFLTGQEEIDTSCEILYERSKMLG 747 Query: 399 PEVGELKCIPLYSTLPPNLQQRIFEPPPPKKANGAIGRKVVVSTNIAETSLTIDGVVFVI 578 + EL +P+YS LP +Q RIFEP PP GRKVV++TNIAETSLTIDG+ +V+ Sbjct: 748 DSIPELVILPVYSALPSEIQSRIFEPAPPG------GRKVVIATNIAETSLTIDGIYYVV 801 Query: 579 DPGFAKQKVFNPRIRVESLLVTPISKASXXXXXXXXXXTRPGKCFRLYTEKGYNNEMHEN 758 DPGF KQ F+P++ ++SL+VTPIS+A T PGKC+RLYTE Y NEM + Sbjct: 802 DPGFVKQSCFDPKLGMDSLIVTPISQAQARQRSGRAGRTGPGKCYRLYTESAYRNEMLPS 861 Query: 759 TYPEILRSNLGTVVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLAALDDDGNLTD 938 PEI R NL +L LK +GI+DL++FDFMDPP +T++ AL+ L L+ALDD+G LT Sbjct: 862 PIPEIQRQNLSHTILMLKAMGINDLLNFDFMDPPPAQTMIAALQNLYALSALDDEGLLTP 921 Query: 939 LGSMMAEFPLDPQLAKMVIASCDFNCSNEILSITAMLSVPQCFVRPTEARTAADEAKMGF 1118 LG MA+FP++PQL+K++I S + CS E+LSI AMLSVP + RP E + AD + F Sbjct: 922 LGRKMADFPMEPQLSKVLITSVELGCSEEMLSIIAMLSVPNIWSRpreKQQEADRQRAQF 981 Query: 1119 AHIDGDHLTMLNVYHAFKQNREDSQWCYEHFVNYRSLKAADNVRIQLSRIMDRFNLRRSS 1298 A+ + DHLT+LNVY +K NR WCYEH++ R ++ A++VR QL R+MDR+ R Sbjct: 982 ANPESDHLTLLNVYTTWKMNRCSDNWCYEHYIQARGMRRAEDVRKQLIRLMDRY--RHPV 1039 Query: 1299 TDFSSKDYYVNIRKALVSGFFMQVAHLE-RSGHYLTVKDNQIVHLHPSTCLDHK-PEWVL 1472 K I +AL SG+F VA + G Y T+ +N V++HPS L K EWV+ Sbjct: 1040 VSCGRKREL--ILRALCSGYFTNVAKRDSHEGCYKTIVENAPVYMHPSGVLFGKAAEWVI 1097 Query: 1473 YNEFVLTTKNFIRTVCEVKPEWLVKVAPQYYDMSN 1577 Y+E + T+K ++ TV V P+WLV+VAP ++ +N Sbjct: 1098 YHELIQTSKEYMHTVSTVNPKWLVEVAPTFFKFAN 1132
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 205,496,717
Number of Sequences: 369166
Number of extensions: 4588790
Number of successful extensions: 13941
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 12694
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 13741
length of database: 68,354,980
effective HSP length: 116
effective length of database: 46,925,720
effective search space used: 20788093960
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail