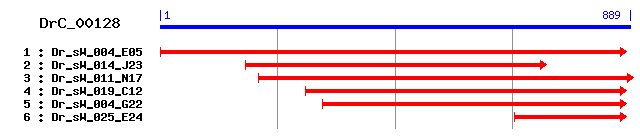
DrC_00128
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00128 (887 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P24156|L2CC_DROME L(2)37Cc protein 276 4e-74 sp|P67778|PHB_MOUSE Prohibitin (B-cell receptor associated ... 261 2e-69 sp|P35232|PHB_HUMAN Prohibitin 259 7e-69 sp|Q9BKU4|PHB1_CAEEL Mitochondrial prohibitin complex prote... 248 2e-65 sp|P40961|PHB_YEAST Prohibitin 225 1e-58 sp|O35129|PHB2_MOUSE Prohibitin-2 (B-cell receptor-associat... 187 4e-47 sp|Q5XIH7|PHB2_RAT Prohibitin-2 (B-cell receptor-associated... 187 4e-47 sp|P50085|PHB2_YEAST Prohibitin-2 185 2e-46 sp|P50093|PHB2_CAEEL Mitochondrial prohibitin complex prote... 176 1e-43 sp|O28852|Y1420_ARCFU Hypothetical protein AF1420 50 8e-06
>sp|P24156|L2CC_DROME L(2)37Cc protein Length = 276 Score = 276 bits (707), Expect = 4e-74 Identities = 137/217 (63%), Positives = 172/217 (79%) Frame = +1 Query: 184 FFIPWIQRPIIYDIRTRPRNIPVNTGSKDLQKVNITLRILYKPVAEMLPKIYTNLGLDFE 363 FFIPW+QRPII+DIR++PRN+PV TGSKDLQ VNITLRILY+P+ + LPKIYT LG D++ Sbjct: 56 FFIPWVQRPIIFDIRSQPRNVPVITGSKDLQNVNITLRILYRPIPDQLPKIYTILGQDYD 115 Query: 364 ERVLPSITYEVLKTVVAQFDASDLITQRELVSQKVNEMLTTRADNFGLTLDDISLTHLTF 543 ERVLPSI EVLK VVAQFDA +LITQRE+VSQ+V++ LT RA FG LDDISLTHLTF Sbjct: 116 ERVLPSIAPEVLKAVVAQFDAGELITQREMVSQRVSQELTVRAKQFGFILDDISLTHLTF 175 Query: 544 GSEFSXXXXXXXXXXXXXXXXXYLVEKAEQEKLAAIISAEGDSQAALLLAKSFNQCGEGF 723 G EF+ ++VEKAEQ+KLA+IISAEGD++AA LLAKSF + G+G Sbjct: 176 GREFTLAVEMKQVAQQEAEKARFVVEKAEQQKLASIISAEGDAEAAGLLAKSFGEAGDGL 235 Query: 724 IELQRIQTSEEIAHLLSKNRNIVYLPDKQGTLINLPA 834 +EL+RI+ +E+IA+ LS++R + YLP Q TL+NLP+ Sbjct: 236 VELRRIEAAEDIAYQLSRSRGVAYLPSGQSTLLNLPS 272
Score = 51.6 bits (122), Expect = 3e-06
Identities = 21/29 (72%), Positives = 25/29 (86%)
Frame = +3
Query: 99 YTVPGGHRAVIFDRISGVRQEVVGEGTHF 185
Y V GGHRAVIFDR +G+++ VVGEGTHF
Sbjct: 28 YNVEGGHRAVIFDRFTGIKENVVGEGTHF 56
>sp|P67778|PHB_MOUSE Prohibitin (B-cell receptor associated protein 32) (BAP 32) sp|P67779|PHB_RAT Prohibitin Length = 272 Score = 261 bits (666), Expect = 2e-69 Identities = 130/216 (60%), Positives = 168/216 (77%) Frame = +1 Query: 184 FFIPWIQRPIIYDIRTRPRNIPVNTGSKDLQKVNITLRILYKPVAEMLPKIYTNLGLDFE 363 F IPW+Q+PII+D R+RPRN+PV TGSKDLQ VNITLRIL++PVA LP+IYT++G D++ Sbjct: 56 FLIPWVQKPIIFDCRSRPRNVPVITGSKDLQNVNITLRILFRPVASQLPRIYTSIGEDYD 115 Query: 364 ERVLPSITYEVLKTVVAQFDASDLITQRELVSQKVNEMLTTRADNFGLTLDDISLTHLTF 543 ERVLPSIT E+LK+VVA+FDA +LITQRELVS++V++ LT RA FGL LDD+SLTHLTF Sbjct: 116 ERVLPSITTEILKSVVARFDAGELITQRELVSRQVSDDLTERAATFGLILDDVSLTHLTF 175 Query: 544 GSEFSXXXXXXXXXXXXXXXXXYLVEKAEQEKLAAIISAEGDSQAALLLAKSFNQCGEGF 723 G EF+ ++VEKAEQ+K AAIISAEGDS+AA L+A S G+G Sbjct: 176 GKEFTEAVEAKQVAQQEAERARFVVEKAEQQKKAAIISAEGDSKAAELIANSLATAGDGL 235 Query: 724 IELQRIQTSEEIAHLLSKNRNIVYLPDKQGTLINLP 831 IEL++++ +E+IA+ LS++RNI YLP Q L+ LP Sbjct: 236 IELRKLEAAEDIAYQLSRSRNITYLPAGQSVLLQLP 271
Score = 48.1 bits (113), Expect = 3e-05
Identities = 22/30 (73%), Positives = 23/30 (76%)
Frame = +3
Query: 99 YTVPGGHRAVIFDRISGVRQEVVGEGTHFL 188
Y V GHRAVIFDR GV+ VVGEGTHFL
Sbjct: 28 YNVDAGHRAVIFDRFRGVQDIVVGEGTHFL 57
>sp|P35232|PHB_HUMAN Prohibitin Length = 272 Score = 259 bits (662), Expect = 7e-69 Identities = 129/216 (59%), Positives = 168/216 (77%) Frame = +1 Query: 184 FFIPWIQRPIIYDIRTRPRNIPVNTGSKDLQKVNITLRILYKPVAEMLPKIYTNLGLDFE 363 F IPW+Q+PII+D R+RPRN+PV TGSKDLQ VNITLRIL++PVA LP+I+T++G D++ Sbjct: 56 FLIPWVQKPIIFDCRSRPRNVPVITGSKDLQNVNITLRILFRPVASQLPRIFTSIGEDYD 115 Query: 364 ERVLPSITYEVLKTVVAQFDASDLITQRELVSQKVNEMLTTRADNFGLTLDDISLTHLTF 543 ERVLPSIT E+LK+VVA+FDA +LITQRELVS++V++ LT RA FGL LDD+SLTHLTF Sbjct: 116 ERVLPSITTEILKSVVARFDAGELITQRELVSRQVSDDLTERAATFGLILDDVSLTHLTF 175 Query: 544 GSEFSXXXXXXXXXXXXXXXXXYLVEKAEQEKLAAIISAEGDSQAALLLAKSFNQCGEGF 723 G EF+ ++VEKAEQ+K AAIISAEGDS+AA L+A S G+G Sbjct: 176 GKEFTEAVEAKQVAQQEAERARFVVEKAEQQKKAAIISAEGDSKAAELIANSLATAGDGL 235 Query: 724 IELQRIQTSEEIAHLLSKNRNIVYLPDKQGTLINLP 831 IEL++++ +E+IA+ LS++RNI YLP Q L+ LP Sbjct: 236 IELRKLEAAEDIAYQLSRSRNITYLPAGQSVLLQLP 271
Score = 48.1 bits (113), Expect = 3e-05
Identities = 22/30 (73%), Positives = 23/30 (76%)
Frame = +3
Query: 99 YTVPGGHRAVIFDRISGVRQEVVGEGTHFL 188
Y V GHRAVIFDR GV+ VVGEGTHFL
Sbjct: 28 YNVDAGHRAVIFDRFRGVQDIVVGEGTHFL 57
>sp|Q9BKU4|PHB1_CAEEL Mitochondrial prohibitin complex protein 1 (Prohibitin-1) Length = 275 Score = 248 bits (633), Expect = 2e-65 Identities = 124/215 (57%), Positives = 158/215 (73%) Frame = +1 Query: 184 FFIPWIQRPIIYDIRTRPRNIPVNTGSKDLQKVNITLRILYKPVAEMLPKIYTNLGLDFE 363 F IPW+Q+PII+DIR+ PR + TGSKDLQ VNITLRIL++P + LP IY N+GLD+ Sbjct: 59 FLIPWVQKPIIFDIRSTPRAVTTITGSKDLQNVNITLRILHRPSPDRLPNIYLNIGLDYA 118 Query: 364 ERVLPSITYEVLKTVVAQFDASDLITQRELVSQKVNEMLTTRADNFGLTLDDISLTHLTF 543 ERVLPSIT EVLK VVAQFDA ++ITQRE+VSQ+ + L RA FGL LDDI++THL F Sbjct: 119 ERVLPSITNEVLKAVVAQFDAHEMITQREVVSQRASVALRERAAQFGLLLDDIAITHLNF 178 Query: 544 GSEFSXXXXXXXXXXXXXXXXXYLVEKAEQEKLAAIISAEGDSQAALLLAKSFNQCGEGF 723 G EF+ YLVEKAEQ K+AA+ +AEGD+QAA LLAK+F G+G Sbjct: 179 GREFTEAVEMKQVAQQEAEKARYLVEKAEQMKIAAVTTAEGDAQAAKLLAKAFASAGDGL 238 Query: 724 IELQRIQTSEEIAHLLSKNRNIVYLPDKQGTLINL 828 +EL++I+ +EEIA ++KN+N+ YLP Q TL+NL Sbjct: 239 VELRKIEAAEEIAERMAKNKNVTYLPGNQQTLLNL 273
Score = 52.8 bits (125), Expect = 1e-06
Identities = 24/30 (80%), Positives = 25/30 (83%)
Frame = +3
Query: 99 YTVPGGHRAVIFDRISGVRQEVVGEGTHFL 188
Y V GG RAVIFDR SGV+ EVVGEGTHFL
Sbjct: 31 YNVDGGQRAVIFDRFSGVKNEVVGEGTHFL 60
>sp|P40961|PHB_YEAST Prohibitin Length = 287 Score = 225 bits (574), Expect = 1e-58 Identities = 103/208 (49%), Positives = 158/208 (75%) Frame = +1 Query: 184 FFIPWIQRPIIYDIRTRPRNIPVNTGSKDLQKVNITLRILYKPVAEMLPKIYTNLGLDFE 363 F +PW+Q+ IIYD+RT+P++I NTG+KDLQ V++TLR+L++P LP IY NLGLD++ Sbjct: 58 FLVPWLQKAIIYDVRTKPKSIATNTGTKDLQMVSLTLRVLHRPEVLQLPAIYQNLGLDYD 117 Query: 364 ERVLPSITYEVLKTVVAQFDASDLITQRELVSQKVNEMLTTRADNFGLTLDDISLTHLTF 543 ERVLPSI EVLK++VAQFDA++LITQRE++SQK+ + L+TRA+ FG+ L+D+S+TH+TF Sbjct: 118 ERVLPSIGNEVLKSIVAQFDAAELITQREIISQKIRKELSTRANEFGIKLEDVSITHMTF 177 Query: 544 GSEFSXXXXXXXXXXXXXXXXXYLVEKAEQEKLAAIISAEGDSQAALLLAKSFNQCGEGF 723 G EF+ +LVEKAEQE+ A++I AEG++++A ++K+ + G+G Sbjct: 178 GPEFTKAVEQKQIAQQDAERAKFLVEKAEQERQASVIRAEGEAESAEFISKALAKVGDGL 237 Query: 724 IELQRIQTSEEIAHLLSKNRNIVYLPDK 807 + ++R++ S++IA L+ + N+VYLP + Sbjct: 238 LLIRRLEASKDIAQTLANSSNVVYLPSQ 265
Score = 52.4 bits (124), Expect = 2e-06
Identities = 25/37 (67%), Positives = 28/37 (75%)
Frame = +3
Query: 99 YTVPGGHRAVIFDRISGVRQEVVGEGTHFLYTMDSKA 209
Y V GG R VIFDRI+GV+Q+VVGEGTHFL KA
Sbjct: 30 YDVKGGSRGVIFDRINGVKQQVVGEGTHFLVPWLQKA 66
>sp|O35129|PHB2_MOUSE Prohibitin-2 (B-cell receptor-associated protein BAP37) (Repressor of estrogen receptor activity) sp|Q99623|PHB2_HUMAN Prohibitin-2 (B-cell receptor-associated protein BAP37) (Repressor of estrogen receptor activity) (D-prohibitin) Length = 299 Score = 187 bits (474), Expect = 4e-47 Identities = 98/205 (47%), Positives = 140/205 (68%) Frame = +1 Query: 184 FFIPWIQRPIIYDIRTRPRNIPVNTGSKDLQKVNITLRILYKPVAEMLPKIYTNLGLDFE 363 F IPW Q PIIYDIR RPR I TGSKDLQ VNI+LR+L +P A+ LP +Y LGLD+E Sbjct: 70 FRIPWFQYPIIYDIRARPRKISSPTGSKDLQMVNISLRVLSRPNAQELPSMYQRLGLDYE 129 Query: 364 ERVLPSITYEVLKTVVAQFDASDLITQRELVSQKVNEMLTTRADNFGLTLDDISLTHLTF 543 ERVLPSI EVLK+VVA+F+AS LITQR VS + LT RA +F L LDD+++T L+F Sbjct: 130 ERVLPSIVNEVLKSVVAKFNASQLITQRAQVSLLIRRELTERAKDFSLILDDVAITELSF 189 Query: 544 GSEFSXXXXXXXXXXXXXXXXXYLVEKAEQEKLAAIISAEGDSQAALLLAKSFNQCGEGF 723 E++ +LVEKA+QE+ I+ AEG+++AA +L ++ ++ G+ Sbjct: 190 SREYTAAVEAKQVAQQEAQRAQFLVEKAKQEQRQKIVQAEGEAEAAKMLGEALSK-NPGY 248 Query: 724 IELQRIQTSEEIAHLLSKNRNIVYL 798 I+L++I+ ++ I+ ++ ++N +YL Sbjct: 249 IKLRKIRAAQNISKTIATSQNRIYL 273
Score = 40.4 bits (93), Expect = 0.007
Identities = 17/30 (56%), Positives = 24/30 (80%), Gaps = 1/30 (3%)
Frame = +3
Query: 99 YTVPGGHRAVIFDRISGVRQE-VVGEGTHF 185
+TV GGHRA+ F+RI GV+Q+ ++ EG HF
Sbjct: 41 FTVEGGHRAIFFNRIGGVQQDTILAEGLHF 70
>sp|Q5XIH7|PHB2_RAT Prohibitin-2 (B-cell receptor-associated protein BAP37) (BAP-37) Length = 299 Score = 187 bits (474), Expect = 4e-47 Identities = 98/205 (47%), Positives = 140/205 (68%) Frame = +1 Query: 184 FFIPWIQRPIIYDIRTRPRNIPVNTGSKDLQKVNITLRILYKPVAEMLPKIYTNLGLDFE 363 F IPW Q PIIYDIR RPR I TGSKDLQ VNI+LR+L +P A+ LP +Y LGLD+E Sbjct: 70 FRIPWFQYPIIYDIRARPRKISSPTGSKDLQMVNISLRVLSRPNAQELPSMYQRLGLDYE 129 Query: 364 ERVLPSITYEVLKTVVAQFDASDLITQRELVSQKVNEMLTTRADNFGLTLDDISLTHLTF 543 ERVLPSI EVLK+VVA+F+AS LITQR VS + LT RA +F L LDD+++T L+F Sbjct: 130 ERVLPSIVNEVLKSVVAKFNASQLITQRAQVSLLIRRELTERAKDFSLILDDVAITELSF 189 Query: 544 GSEFSXXXXXXXXXXXXXXXXXYLVEKAEQEKLAAIISAEGDSQAALLLAKSFNQCGEGF 723 E++ +LVEKA+QE+ I+ AEG+++AA +L ++ ++ G+ Sbjct: 190 SREYTAAVEAKQVAQQEAQRAQFLVEKAKQEQRQKIVQAEGEAEAAKMLGEALSK-NPGY 248 Query: 724 IELQRIQTSEEIAHLLSKNRNIVYL 798 I+L++I+ ++ I+ ++ ++N +YL Sbjct: 249 IKLRKIRAAQNISKTIATSQNRIYL 273
Score = 40.4 bits (93), Expect = 0.007
Identities = 17/30 (56%), Positives = 24/30 (80%), Gaps = 1/30 (3%)
Frame = +3
Query: 99 YTVPGGHRAVIFDRISGVRQE-VVGEGTHF 185
+TV GGHRA+ F+RI GV+Q+ ++ EG HF
Sbjct: 41 FTVEGGHRAIFFNRIGGVQQDTILAEGLHF 70
>sp|P50085|PHB2_YEAST Prohibitin-2 Length = 315 Score = 185 bits (469), Expect = 2e-46 Identities = 95/214 (44%), Positives = 138/214 (64%) Frame = +1 Query: 184 FFIPWIQRPIIYDIRTRPRNIPVNTGSKDLQKVNITLRILYKPVAEMLPKIYTNLGLDFE 363 F PW+ PIIYD+R +PRN+ TG+KDLQ VNIT R+L +P LP IY LG D++ Sbjct: 87 FIFPWLDTPIIYDVRAKPRNVASLTGTKDLQMVNITCRVLSRPDVVQLPTIYRTLGQDYD 146 Query: 364 ERVLPSITYEVLKTVVAQFDASDLITQRELVSQKVNEMLTTRADNFGLTLDDISLTHLTF 543 ERVLPSI EVLK VVAQF+AS LITQRE VS+ + E L RA F + LDD+S+T++TF Sbjct: 147 ERVLPSIVNEVLKAVVAQFNASQLITQREKVSRLIRENLVRRASKFNILLDDVSITYMTF 206 Query: 544 GSEFSXXXXXXXXXXXXXXXXXYLVEKAEQEKLAAIISAEGDSQAALLLAKSFNQCGEGF 723 EF+ ++V+KA QEK ++ A+G++++A L+ ++ + + Sbjct: 207 SPEFTNAVEAKQIAQQDAQRAAFVVDKARQEKQGMVVRAQGEAKSAELIGEAIKK-SRDY 265 Query: 724 IELQRIQTSEEIAHLLSKNRNIVYLPDKQGTLIN 825 +EL+R+ T+ +IA +L+ + N V L D + L+N Sbjct: 266 VELKRLDTARDIAKILASSPNRVIL-DNEALLLN 298
Score = 42.4 bits (98), Expect = 0.002
Identities = 15/31 (48%), Positives = 22/31 (70%)
Frame = +3
Query: 99 YTVPGGHRAVIFDRISGVRQEVVGEGTHFLY 191
+ V GGHRA+++ RI GV + EGTHF++
Sbjct: 59 FNVDGGHRAIVYSRIHGVSSRIFNEGTHFIF 89
>sp|P50093|PHB2_CAEEL Mitochondrial prohibitin complex protein 2 (Prohibitin-2) Length = 286 Score = 176 bits (445), Expect = 1e-43 Identities = 99/215 (46%), Positives = 138/215 (64%) Frame = +1 Query: 184 FFIPWIQRPIIYDIRTRPRNIPVNTGSKDLQKVNITLRILYKPVAEMLPKIYTNLGLDFE 363 F IPW Q PIIYDIR RP I TGSKDLQ VNI LR+L +P E L IY LG ++E Sbjct: 61 FRIPWFQYPIIYDIRARPNQIRSPTGSKDLQMVNIGLRVLSRPNPEHLVHIYRTLGQNWE 120 Query: 364 ERVLPSITYEVLKTVVAQFDASDLITQRELVSQKVNEMLTTRADNFGLTLDDISLTHLTF 543 ERVLPSI EVLK VVA+F+AS LITQR+ VS V + L RA +F + LDD+SLT L F Sbjct: 121 ERVLPSICNEVLKGVVAKFNASQLITQRQQVSMLVRKTLIERALDFNIILDDVSLTELAF 180 Query: 544 GSEFSXXXXXXXXXXXXXXXXXYLVEKAEQEKLAAIISAEGDSQAALLLAKSFNQCGEGF 723 ++S + VE+A+Q+K I+ AEG++++A LL ++ GF Sbjct: 181 SPQYSAAVEAKQVAAQEAQRATFYVERAKQQKQEKIVQAEGEAESAKLLGEAMKN-DPGF 239 Query: 724 IELQRIQTSEEIAHLLSKNRNIVYLPDKQGTLINL 828 ++L++I+ +++IA ++S++ N YLP G ++N+ Sbjct: 240 LKLRKIRAAQKIARIVSESGNKTYLP-TGGLMLNI 273
Score = 36.6 bits (83), Expect = 0.096
Identities = 14/29 (48%), Positives = 21/29 (72%)
Frame = +3
Query: 99 YTVPGGHRAVIFDRISGVRQEVVGEGTHF 185
+TV GHRA++F+RI G+ ++ EG HF
Sbjct: 33 FTVEAGHRAIMFNRIGGLSTDLYKEGLHF 61
>sp|O28852|Y1420_ARCFU Hypothetical protein AF1420 Length = 249 Score = 50.1 bits (118), Expect = 8e-06 Identities = 40/198 (20%), Positives = 94/198 (47%), Gaps = 5/198 (2%) Frame = +1 Query: 184 FFIPWIQRPIIYDIRTRPRNIPVNTGSKDLQKVNITLRI---LYKPVAEMLPKIYTNLGL 354 F IP ++ ++ D+RT ++P + + K N+T+++ +Y V + + Sbjct: 47 FIIPILENMVVVDLRTVTYDVP---SQEVVTKDNVTVKVNAVVYYRVVDPAKAVTEVFDY 103 Query: 355 DFEERVLPSITYEVLKTVVAQFDASDLITQRELVSQKVNEMLTTRADNFGLTLDDISLTH 534 + L T L++++ Q + +++++R+ ++ K+ +++ + +G+ + + + Sbjct: 104 QYATAQLAQTT---LRSIIGQAELDEVLSERDKLNVKLQQIIDEETNPWGIKVTAVEIKD 160 Query: 535 LTFGSEFSXXXXXXXXXXXXXXXXXYLVEKAEQEKLAAIISAEGDSQAALLLAKSFNQC- 711 + E + +AE+E+ + II AEG+ QAA+ L ++ + Sbjct: 161 VELPEEMRRIMA--------------MQAEAERERRSKIIRAEGEYQAAMKLREAADVLA 206 Query: 712 -GEGFIELQRIQTSEEIA 762 EG I L+ +QT EI+ Sbjct: 207 QSEGAILLRYLQTLNEIS 224
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 93,366,899
Number of Sequences: 369166
Number of extensions: 1754429
Number of successful extensions: 4172
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 4051
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4156
length of database: 68,354,980
effective HSP length: 110
effective length of database: 48,034,130
effective search space used: 8886314050
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail