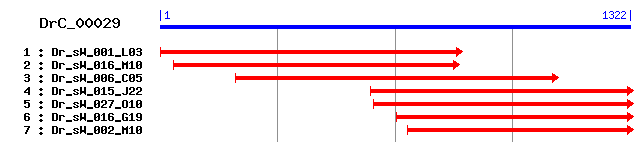
DrC_00029
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00029 (1321 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P32369|BAIG_EUBSP Bile acid transporter 51 7e-06 sp|P37643|YHJE_ECOLI Inner membrane metabolite transport pr... 50 1e-05 sp|P46333|CSBC_BACSU Probable metabolite transport protein ... 50 1e-05 sp|Q92EE1|LMRB_LISIN Lincomycin resistance protein lmrB 49 2e-05 sp|Q8Y9K8|LMRB_LISMO Lincomycin resistance protein lmrB 49 2e-05 sp|Q58955|YF60_METJA Putative transporter MJ1560 49 2e-05 sp|P47234|LACY_CITFR Lactose permease (Lactose-proton symport) 47 1e-04 sp|P46907|NARK_BACSU Nitrite extrusion protein (Nitrite fac... 47 2e-04 sp|P58120|PMRA_LACLA Multi-drug resistance efflux pump pmrA... 42 0.003 sp|P25744|MDTG_ECOLI Multidrug resistance protein mdtG >gi|... 42 0.004
>sp|P32369|BAIG_EUBSP Bile acid transporter Length = 477 Score = 51.2 bits (121), Expect = 7e-06 Identities = 33/144 (22%), Positives = 67/144 (46%), Gaps = 1/144 (0%) Frame = +3 Query: 666 GLNGADAALITSLLYIVSAACSPVMGFLIDLTGRNLFWVLTGTLATLGS-HALYAFTFAN 842 G++ + I+ L + SAAC+P++G L D+ GR +L + G+ A + Sbjct: 42 GIDYNNTTWISLALAMSSAACAPILGKLGDVLGRRTTLLLGIVIFAAGNVLTAVATSLIF 101 Query: 843 PLGVMIFLGVSYSVLASSLWPMVALVIPAHQRGTAFGIMQSVQNLGLAVVAIIAGLIQDK 1022 L +G+ + ++ + + P + G AFG+ + + + V GLI + Sbjct: 102 MLAARFIVGIGTAAISPIVMAYIVTEYPQEETGKAFGLYMLISSGAVVVGPTCGGLIMNA 161 Query: 1023 YGFLILEVFFSVWLCITLVLIVLL 1094 G+ ++ +W+C+ L ++V L Sbjct: 162 AGWRVM-----MWVCVALCVVVFL 180
>sp|P37643|YHJE_ECOLI Inner membrane metabolite transport protein yhjE Length = 440 Score = 50.4 bits (119), Expect = 1e-05 Identities = 53/212 (25%), Positives = 89/212 (41%), Gaps = 13/212 (6%) Frame = +3 Query: 501 ILQRNKKNSEDEKISLGDI--RHFSVAF---WIISGICVAYYVAVFPFVSLSKVFLIQKY 665 + ++ K + KI LG + +H V +I+ +Y+ ++ S Sbjct: 224 VFEKVAKAKKQVKIPLGTLLTKHVRVTVLGTFIMLATYTLFYIMTVYSMTFSTAAAPVGL 283 Query: 666 GLNGADAALITSLLYIVSAACSPVMGFLIDLTGRNLFWVLTGTLATLGSHALYAFT---- 833 GL + + + I PV G L D GR V+ TL L AL+AF Sbjct: 284 GLPRNEVLWMLMMAVIGFGVMVPVAGLLADAFGRRKSMVIITTLIIL--FALFAFNPLLG 341 Query: 834 FANPLGVMIFLGVSYSVLASSLWPMVALV---IPAHQRGTAFGIMQSVQN-LGLAVVAII 1001 NP+ V FL + S++ + PM AL+ P R T +V + LG +V I Sbjct: 342 SGNPILVFAFLLLGLSLMGLTFGPMGALLPELFPTEVRYTGASFSYNVASILGASVAPYI 401 Query: 1002 AGLIQDKYGFLILEVFFSVWLCITLVLIVLLY 1097 A +Q YG + ++ + +TL+ ++L + Sbjct: 402 AAWLQTNYGLGAVGLYLAAMAGLTLIALLLTH 433
>sp|P46333|CSBC_BACSU Probable metabolite transport protein csbC Length = 461 Score = 50.4 bits (119), Expect = 1e-05 Identities = 63/256 (24%), Positives = 110/256 (42%), Gaps = 23/256 (8%) Frame = +3 Query: 501 ILQRNKKNSEDEKISLGDIRHFSVAFWIISGICVAYYVAVFP-FVSLSKVF-----LIQK 662 + + + +E ++ +LG ++ A WI + + +A+F V ++ V + K Sbjct: 216 LAEMKQGEAEKKETTLGVLK----AKWIRPMLLIGVGLAIFQQAVGINTVIYYAPTIFTK 271 Query: 663 YGLNGADAALITSLLYIVSAACSPVMGFLIDLTGRN--LFW----------VLTGTLATL 806 GL + +AL T + I++ LID GR L W L+G L TL Sbjct: 272 AGLGTSASALGTMGIGILNVIMCITAMILIDRVGRKKLLIWGSVGITLSLAALSGVLLTL 331 Query: 807 GSHALYAFTFANPLGVMIFLGVSYSVLASSLWPMVALVI-----PAHQRGTAFGIMQSVQ 971 G A A+ ++FLGV Y V + W V V+ P+ RG A G V Sbjct: 332 GLSASTAWM------TVVFLGV-YIVFYQATWGPVVWVLMPELFPSKARGAATGFTTLVL 384 Query: 972 NLGLAVVAIIAGLIQDKYGFLILEVFFSVWLCITLVLIVLLYLWDVQNGGKLNESGPSRR 1151 + +V+++ L+ G + + FSV +C+ L Y+ G L E S + Sbjct: 385 SAANLIVSLVFPLMLSAMGIAWVFMVFSV-ICL-LSFFFAFYMVPETKGKSLEEIEASLK 442 Query: 1152 KKLKKSEYNVSETMDK 1199 K+ KK + ++ +++ Sbjct: 443 KRFKKKKSTQNQVLNE 458
>sp|Q92EE1|LMRB_LISIN Lincomycin resistance protein lmrB Length = 471 Score = 49.3 bits (116), Expect = 2e-05 Identities = 45/180 (25%), Positives = 83/180 (46%), Gaps = 8/180 (4%) Frame = +3 Query: 582 IISGICVAYYVAVFPFVSLSKVF--LIQKYGLNGADAALITSLLYIVSAACSPVMGFLID 755 II+ +A ++ +F +L+ LIQ + ++ A +T+ + P+ G L+ Sbjct: 16 IIASFLMAGFIGLFSETALNMALSDLIQVFDISSATVQWLTTGYLLTLGILVPISGLLLQ 75 Query: 756 -LTGRNLFWV-----LTGTLATLGSHALYAFTFANPLGVMIFLGVSYSVLASSLWPMVAL 917 T R LF+ + GTL A + TFA + + V ++L ++ + L Sbjct: 76 WFTTRGLFFTAVSFSIAGTLI-----AALSPTFAMLMIGRVVQAVGTALLLPLMFNTILL 130 Query: 918 VIPAHQRGTAFGIMQSVQNLGLAVVAIIAGLIQDKYGFLILEVFFSVWLCITLVLIVLLY 1097 + P H+RG+A G++ V AV I+GLI + L + W+ + ++I LL+ Sbjct: 131 IFPEHKRGSAMGMIGLVIMFAPAVGPTISGLILEN-----LTWNWIFWISLPFLIIALLF 185
>sp|Q8Y9K8|LMRB_LISMO Lincomycin resistance protein lmrB Length = 471 Score = 49.3 bits (116), Expect = 2e-05 Identities = 45/180 (25%), Positives = 83/180 (46%), Gaps = 8/180 (4%) Frame = +3 Query: 582 IISGICVAYYVAVFPFVSLSKVF--LIQKYGLNGADAALITSLLYIVSAACSPVMGFLID 755 II+ +A ++ +F +L+ LIQ + ++ A +T+ + P+ G L+ Sbjct: 16 IIASFLMAGFIGLFSETALNMALSDLIQVFDISSATVQWLTTGYLLTLGILVPISGLLLQ 75 Query: 756 -LTGRNLFWV-----LTGTLATLGSHALYAFTFANPLGVMIFLGVSYSVLASSLWPMVAL 917 T R LF+ + GTL A + TFA + + V ++L ++ + L Sbjct: 76 WFTTRGLFFTAVSFSIAGTLI-----AALSPTFAMLMIGRVVQAVGTALLLPLMFNTILL 130 Query: 918 VIPAHQRGTAFGIMQSVQNLGLAVVAIIAGLIQDKYGFLILEVFFSVWLCITLVLIVLLY 1097 + P H+RG+A G++ V AV I+GLI + L + W+ + ++I LL+ Sbjct: 131 IFPEHKRGSAMGMIGLVIMFAPAVGPTISGLILEN-----LTWNWIFWISLPFLIIALLF 185
>sp|Q58955|YF60_METJA Putative transporter MJ1560 Length = 386 Score = 49.3 bits (116), Expect = 2e-05 Identities = 49/227 (21%), Positives = 106/227 (46%), Gaps = 7/227 (3%) Frame = +3 Query: 441 CVFSLICALVLAMFDRRAERILQRNKKNSEDEKIS------LGDIRHFSVAFWIISGICV 602 C F I A +++ + + + + +NK+ + +KIS R+FS +F II+ V Sbjct: 166 CGFLGILAAIIS-YMKLEDIVFNKNKEKIDVKKISTLFSFEFLKNRNFSSSF-IINVSNV 223 Query: 603 AYYVAVFPFVSLSKVFLIQKYGLNGADAALITSLLYIVSAACSPVMGFLIDLTGRNLFWV 782 ++ +++L + Y + + + +L I+ A G L D G N+ + Sbjct: 224 MINAGIYAYLALYAI----NYNITISQVGFMIALTNILMALLQRSFGKLYDKLG-NIMII 278 Query: 783 LTGTLATLGSHALY-AFTFANPLGVMIFLGVSYSVLASSLWPMVALVIPAHQRGTAFGIM 959 + + + G + L + TF L + + V S+ +++ + IP H++G A G+ Sbjct: 279 IGIFIISFGMYLLSTSTTFLTILASLTIIAVGSSISSTATTSLAVKDIPTHRKGEAMGLF 338 Query: 960 QSVQNLGLAVVAIIAGLIQDKYGFLILEVFFSVWLCITLVLIVLLYL 1100 + N+G+ + A+ G + D G + F +++ ++V+ ++ YL Sbjct: 339 TTSINIGMFIGAVSFGFLADILGIANMYKFSAIF---SIVVGIISYL 382
>sp|P47234|LACY_CITFR Lactose permease (Lactose-proton symport) Length = 416 Score = 47.0 bits (110), Expect = 1e-04 Identities = 45/201 (22%), Positives = 85/201 (42%), Gaps = 14/201 (6%) Frame = +3 Query: 576 FWIISGICVAYYVAVFPFVSLSKVFLIQKYGLNGADAALITSLLYIVSAACSPVMGFLID 755 FW+ Y+ + + ++L + ++ D +I + + + S P+ G L D Sbjct: 9 FWMFGFFFFFYFFIMGAYFPFFPIWLHEVNHISKGDTGIIFACISLFSLLFQPIFGLLSD 68 Query: 756 LTG--RNLFWVLTGTLATLGSHALYAFTFANPLGVM-------IFLGVSYSVLASSLWPM 908 G ++L WV+TG L +Y F + ++ I+LG Y+ A ++ Sbjct: 69 KLGLRKHLLWVITGMLVMFAPFFIYVFGPLLQVNILLGSIVGGIYLGFIYNAGAPAI--- 125 Query: 909 VALVIPAHQRGT-AFGIMQSVQNLGLAVVAIIAGLIQDKYGFLILEVFFSVWL----CIT 1073 A + A +R FG + +G A+ A IAG++ + F WL + Sbjct: 126 EAYIEKASRRSNFEFGRARMFGCVGWALCASIAGIM------FTINNQFVFWLGSGCAVI 179 Query: 1074 LVLIVLLYLWDVQNGGKLNES 1136 L L++L DV + K+ ++ Sbjct: 180 LALLLLFSKTDVPSSAKVADA 200
>sp|P46907|NARK_BACSU Nitrite extrusion protein (Nitrite facilitator) Length = 395 Score = 46.6 bits (109), Expect = 2e-04 Identities = 51/183 (27%), Positives = 80/183 (43%), Gaps = 10/183 (5%) Frame = +3 Query: 579 WIISGICVAYYVAVFPFVSLSKVFLIQKYGLNGADAALITSLLYIVSAACSPVMGFLIDL 758 W +S + A F FL++ +GLN ADA L T+ VS P GFL D Sbjct: 205 WFLSLFYFITFGAFVAFTIYLPNFLVEHFGLNPADAGLRTAGFIAVSTLLRPAGGFLADK 264 Query: 759 TG--RNLFWVLTGTLATLGSHALYAFTFANPLGVMIFLGVSYSVLASSLWPMVALVIP-- 926 R L +V TG TL L +F+ +G+ F ++ +V + V ++P Sbjct: 265 MSPLRILMFVFTG--LTLSGIIL---SFSPTIGLYTFGSLTVAVCSGIGNGTVFKLVPFY 319 Query: 927 -AHQRGTAFGIMQSVQNLG-----LAVVAIIAGLIQDKYGFLILEVFFSVWLCITLVLIV 1088 + Q G A GI+ ++ LG L + ++ Q GF+ L S + VL++ Sbjct: 320 FSKQAGIANGIVSAMGGLGGFFPPLILASVFQATGQYAIGFMAL----SEVALASFVLVI 375 Query: 1089 LLY 1097 +Y Sbjct: 376 WMY 378
>sp|P58120|PMRA_LACLA Multi-drug resistance efflux pump pmrA homolog Length = 405 Score = 42.4 bits (98), Expect = 0.003 Identities = 42/178 (23%), Positives = 76/178 (42%), Gaps = 5/178 (2%) Frame = +3 Query: 579 WIISGICVAYYVAVFPFVSLSKVFLIQKYGLNGADAALITSLLYIVSAACS----PVMGF 746 WI + + V PF+ L IQ G++G + L + L + + A S P+ G Sbjct: 17 WIGCFFVGSSFSLVMPFLPL----YIQGLGVSGGNVELYSGLAFSLPALASGLVAPIWGR 72 Query: 747 LIDLTGRNLFWVLTGTLATLGSHAL-YAFTFANPLGVMIFLGVSYSVLASSLWPMVALVI 923 L D GR + V + TL + +A LG+ + +G + +S M+A Sbjct: 73 LADEHGRKVMMVRASIVMTLTMGGIAFAPNVWWLLGLRLLMGFFSGYIPNST-AMIASQA 131 Query: 924 PAHQRGTAFGIMQSVQNLGLAVVAIIAGLIQDKYGFLILEVFFSVWLCITLVLIVLLY 1097 P + G A G + + G + + GL+ + +G + VF V + L ++ ++ Sbjct: 132 PKDKSGYALGTLATAMVSGTLIGPSLGGLLAEWFG--MANVFLIVGALLALATLLTIF 187
>sp|P25744|MDTG_ECOLI Multidrug resistance protein mdtG sp|Q8X9I3|MDTG_ECO57 Multidrug resistance protein mdtG Length = 408 Score = 42.0 bits (97), Expect = 0.004 Identities = 45/185 (24%), Positives = 80/185 (43%), Gaps = 9/185 (4%) Frame = +3 Query: 570 VAFWIISGICVAYYVAVFPFVSLSKVFLIQKYGLNGADA-----ALITSLLYIVSAACSP 734 + W+ + A + V PF+ L +++ G+ G A ++ S+ ++ SA SP Sbjct: 17 IVAWLGCFLTGAAFSLVMPFLPL----YVEQLGVTGHSALNMWSGIVFSITFLFSAIASP 72 Query: 735 VMGFLIDLTGRNLFWVLT----GTLATLGSHALYAFTFANPLGVMIFLGVSYSVLASSLW 902 G L D GR L + + G + L A + F ++ LG + A++L Sbjct: 73 FWGGLADRKGRKLMLLRSALGMGIVMVLMGLAQNIWQFLILRALLGLLG-GFVPNANAL- 130 Query: 903 PMVALVIPAHQRGTAFGIMQSVQNLGLAVVAIIAGLIQDKYGFLILEVFFSVWLCITLVL 1082 +A +P ++ G A G + + G + + GL+ D YG + VFF + L Sbjct: 131 --IATQVPRNKSGWALGTLSTGGVSGALLGPMAGGLLADSYG--LRPVFFITASVLILCF 186 Query: 1083 IVLLY 1097 V L+ Sbjct: 187 FVTLF 191
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 147,528,254
Number of Sequences: 369166
Number of extensions: 3014744
Number of successful extensions: 8577
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8095
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8555
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 15370936750
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail