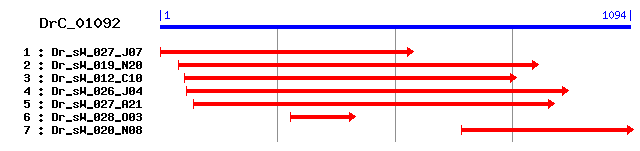
DrC_01092
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01092 (1094 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P63017|HSP7C_MOUSE Heat shock cognate 71 kDa protein (He... 449 e-126 sp|P19378|HSP7C_CRIGR Heat shock cognate 71 kDa protein (He... 449 e-126 sp|P11142|HSP7C_HUMAN Heat shock cognate 71 kDa protein (He... 448 e-125 sp|Q90473|HSP7C_BRARE Heat shock cognate 71 kDa protein (He... 447 e-125 sp|P09446|HSP7A_CAEEL Heat shock 70 kDa protein A 446 e-125 sp|P19120|HSP7C_BOVIN Heat shock cognate 71 kDa protein (He... 445 e-125 sp|P08108|HSP70_ONCMY Heat shock cognate 70 kDa protein (HS... 444 e-124 sp|P08106|HSP70_CHICK Heat shock 70 kDa protein (HSP70) 444 e-124 sp|Q9U639|HSP7D_MANSE Heat shock 70 kDa protein cognate 4 (... 443 e-124 sp|Q24789|HSP70_ECHGR Heat shock cognate 70 kDa protein (HS... 442 e-124
>sp|P63017|HSP7C_MOUSE Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8) sp|P63018|HSP7C_RAT Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8) Length = 646 Score = 449 bits (1156), Expect = e-126 Identities = 232/344 (67%), Positives = 262/344 (76%), Gaps = 1/344 (0%) Frame = +3 Query: 15 EELNADLFRSTLEPVEKALRDAKLDKNAIHEIVLVGGSTRIPKIQKLLQDLFNGKELNKS 194 EELNADLFR TL+PVEKALRDAKLDK+ IH+IVLVGGSTRIPKIQKLLQD FNGKELNKS Sbjct: 303 EELNADLFRGTLDPVEKALRDAKLDKSQIHDIVLVGGSTRIPKIQKLLQDFFNGKELNKS 362 Query: 195 INPDEXXXXXXXXXXXILSGDKSEAVQDXXXXXXXXXXXGIETAGGVMTALIKRNTTIPT 374 INPDE ILSGDKSE VQD GIETAGGVMT LIKRNTTIPT Sbjct: 363 INPDEAVAYGAAVQAAILSGDKSENVQDLLLLDVTPLSLGIETAGGVMTVLIKRNTTIPT 422 Query: 375 KQTQIFSTYSDNQPGVLIQVFEGERAMTKDNNLLGKFELSGIPPAPRGVPQIEVTFDIDA 554 KQTQ F+TYSDNQPGVLIQV+EGERAMTKDNNLLGKFEL+GIPPAPRGVPQIEVTFDIDA Sbjct: 423 KQTQTFTTYSDNQPGVLIQVYEGERAMTKDNNLLGKFELTGIPPAPRGVPQIEVTFDIDA 482 Query: 555 NGILNVSATEKGTGKENKITITNDKGRLSKDDIERMVNXXXXXXXXXXXXXXRIGAKNLL 734 NGILNVSA +K TGKENKITITNDKGRLSK+DIERMV ++ +KN L Sbjct: 483 NGILNVSAVDKSTGKENKITITNDKGRLSKEDIERMVQEAEKYKAEDEKQRDKVSSKNSL 542 Query: 735 ESYAYQMKSTIEDEKLKDKISESDRKQIVDRCQEAISWLEKNQQATKEEFDYHYKDLEKL 914 ESYA+ MK+T+EDEKL+ KI++ D+++I+D+C E ISWL+KNQ A KEEF++ K+LEK+ Sbjct: 543 ESYAFNMKATVEDEKLQGKINDEDKQKILDKCNEIISWLDKNQTAEKEEFEHQQKELEKV 602 Query: 915 CNPIMTKMYQ-XXXXXXXXXXXXXXXXXXXXXXKRQGPTVEEVD 1043 CNPI+TK+YQ GPT+EEVD Sbjct: 603 CNPIITKLYQSAGGMPGGMPGGFPGGGAPPSGGASSGPTIEEVD 646
>sp|P19378|HSP7C_CRIGR Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8) Length = 646 Score = 449 bits (1156), Expect = e-126 Identities = 232/344 (67%), Positives = 262/344 (76%), Gaps = 1/344 (0%) Frame = +3 Query: 15 EELNADLFRSTLEPVEKALRDAKLDKNAIHEIVLVGGSTRIPKIQKLLQDLFNGKELNKS 194 EELNADLFR TL+PVEKALRDAKLDK+ IH+IVLVGGSTRIPKIQKLLQD FNGKELNKS Sbjct: 303 EELNADLFRGTLDPVEKALRDAKLDKSQIHDIVLVGGSTRIPKIQKLLQDFFNGKELNKS 362 Query: 195 INPDEXXXXXXXXXXXILSGDKSEAVQDXXXXXXXXXXXGIETAGGVMTALIKRNTTIPT 374 INPDE ILSGDKSE VQD GIETAGGVMT LIKRNTTIPT Sbjct: 363 INPDEAVAYGAAVQAAILSGDKSENVQDLLLLDVTPLSLGIETAGGVMTVLIKRNTTIPT 422 Query: 375 KQTQIFSTYSDNQPGVLIQVFEGERAMTKDNNLLGKFELSGIPPAPRGVPQIEVTFDIDA 554 KQTQ F+TYSDNQPGVLIQV+EGERAMTKDNNLLGKFEL+GIPPAPRGVPQIEVTFDIDA Sbjct: 423 KQTQTFTTYSDNQPGVLIQVYEGERAMTKDNNLLGKFELTGIPPAPRGVPQIEVTFDIDA 482 Query: 555 NGILNVSATEKGTGKENKITITNDKGRLSKDDIERMVNXXXXXXXXXXXXXXRIGAKNLL 734 NGILNVSA +K TGKENKITITNDKGRLSK+DIERMV ++ +KN L Sbjct: 483 NGILNVSAVDKSTGKENKITITNDKGRLSKEDIERMVQEAEKYKAEDEKQRDKVSSKNSL 542 Query: 735 ESYAYQMKSTIEDEKLKDKISESDRKQIVDRCQEAISWLEKNQQATKEEFDYHYKDLEKL 914 ESYA+ MK+T+EDEKL+ KI++ D+++I+D+C E ISWL+KNQ A KEEF++ K+LEK+ Sbjct: 543 ESYAFNMKATVEDEKLQGKINDEDKQKILDKCNEIISWLDKNQTAEKEEFEHQQKELEKV 602 Query: 915 CNPIMTKMYQ-XXXXXXXXXXXXXXXXXXXXXXKRQGPTVEEVD 1043 CNPI+TK+YQ GPT+EEVD Sbjct: 603 CNPIITKLYQSAGGMPGGMPGGFPGGGAPPSGGASSGPTIEEVD 646
>sp|P11142|HSP7C_HUMAN Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8) sp|Q71U34|HSP7C_SAGOE Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8) (Intracellular vitamin D binding protein 1) Length = 646 Score = 448 bits (1153), Expect = e-125 Identities = 231/344 (67%), Positives = 262/344 (76%), Gaps = 1/344 (0%) Frame = +3 Query: 15 EELNADLFRSTLEPVEKALRDAKLDKNAIHEIVLVGGSTRIPKIQKLLQDLFNGKELNKS 194 EELNADLFR TL+PVEKALRDAKLDK+ IH+IVLVGGSTRIPKIQKLLQD FNGKELNKS Sbjct: 303 EELNADLFRGTLDPVEKALRDAKLDKSQIHDIVLVGGSTRIPKIQKLLQDFFNGKELNKS 362 Query: 195 INPDEXXXXXXXXXXXILSGDKSEAVQDXXXXXXXXXXXGIETAGGVMTALIKRNTTIPT 374 INPDE ILSGDKSE VQD GIETAGGVMT LIKRNTTIPT Sbjct: 363 INPDEAVAYGAAVQAAILSGDKSENVQDLLLLDVTPLSLGIETAGGVMTVLIKRNTTIPT 422 Query: 375 KQTQIFSTYSDNQPGVLIQVFEGERAMTKDNNLLGKFELSGIPPAPRGVPQIEVTFDIDA 554 KQTQ F+TYSDNQPGVLIQV+EGERAMTKDNNLLGKFEL+GIPPAPRGVPQIEVTFDIDA Sbjct: 423 KQTQTFTTYSDNQPGVLIQVYEGERAMTKDNNLLGKFELTGIPPAPRGVPQIEVTFDIDA 482 Query: 555 NGILNVSATEKGTGKENKITITNDKGRLSKDDIERMVNXXXXXXXXXXXXXXRIGAKNLL 734 NGILNVSA +K TGKENKITITNDKGRLSK+DIERMV ++ +KN L Sbjct: 483 NGILNVSAVDKSTGKENKITITNDKGRLSKEDIERMVQEAEKYKAEDEKQRDKVSSKNSL 542 Query: 735 ESYAYQMKSTIEDEKLKDKISESDRKQIVDRCQEAISWLEKNQQATKEEFDYHYKDLEKL 914 ESYA+ MK+T+EDEKL+ KI++ D+++I+D+C E I+WL+KNQ A KEEF++ K+LEK+ Sbjct: 543 ESYAFNMKATVEDEKLQGKINDEDKQKILDKCNEIINWLDKNQTAEKEEFEHQQKELEKV 602 Query: 915 CNPIMTKMYQ-XXXXXXXXXXXXXXXXXXXXXXKRQGPTVEEVD 1043 CNPI+TK+YQ GPT+EEVD Sbjct: 603 CNPIITKLYQSAGGMPGGMPGGFPGGGAPPSGGASSGPTIEEVD 646
>sp|Q90473|HSP7C_BRARE Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8) Length = 649 Score = 447 bits (1150), Expect = e-125 Identities = 232/347 (66%), Positives = 260/347 (74%), Gaps = 4/347 (1%) Frame = +3 Query: 15 EELNADLFRSTLEPVEKALRDAKLDKNAIHEIVLVGGSTRIPKIQKLLQDLFNGKELNKS 194 EELNADLFR TL+PVEKALRDAK+DK IH+IVLVGGSTRIPKIQKLLQD FNGKELNKS Sbjct: 303 EELNADLFRGTLDPVEKALRDAKMDKAQIHDIVLVGGSTRIPKIQKLLQDYFNGKELNKS 362 Query: 195 INPDEXXXXXXXXXXXILSGDKSEAVQDXXXXXXXXXXXGIETAGGVMTALIKRNTTIPT 374 INPDE ILSGDKSE VQD GIETAGGVMT LIKRNTTIPT Sbjct: 363 INPDEAVAYGAAVQAAILSGDKSENVQDLLLLDVTPLSLGIETAGGVMTVLIKRNTTIPT 422 Query: 375 KQTQIFSTYSDNQPGVLIQVFEGERAMTKDNNLLGKFELSGIPPAPRGVPQIEVTFDIDA 554 KQTQ F+TYSDNQPGVLIQV+EGERAMTKDNNLLGKFEL+GIPPAPRGVPQIEVTFDIDA Sbjct: 423 KQTQTFTTYSDNQPGVLIQVYEGERAMTKDNNLLGKFELTGIPPAPRGVPQIEVTFDIDA 482 Query: 555 NGILNVSATEKGTGKENKITITNDKGRLSKDDIERMVNXXXXXXXXXXXXXXRIGAKNLL 734 NGI+NVSA +K TGKENKITITNDKGRLSK+DIERMV ++ AKN L Sbjct: 483 NGIMNVSAVDKSTGKENKITITNDKGRLSKEDIERMVQEAEKYKAEDDVQRDKVSAKNGL 542 Query: 735 ESYAYQMKSTIEDEKLKDKISESDRKQIVDRCQEAISWLEKNQQATKEEFDYHYKDLEKL 914 ESYA+ MKST+EDEKLK KIS+ D+++I+D+C E I WL+KNQ A +EEF++ K+LEK+ Sbjct: 543 ESYAFNMKSTVEDEKLKGKISDEDKQKILDKCNEVIGWLDKNQTAEREEFEHQQKELEKV 602 Query: 915 CNPIMTKMYQXXXXXXXXXXXXXXXXXXXXXXK----RQGPTVEEVD 1043 CNPI+TK+YQ GPT+EEVD Sbjct: 603 CNPIITKLYQSAGGMPGGMPEGMPGGFPGAGAAPGGGSSGPTIEEVD 649
>sp|P09446|HSP7A_CAEEL Heat shock 70 kDa protein A Length = 640 Score = 446 bits (1146), Expect = e-125 Identities = 233/343 (67%), Positives = 259/343 (75%) Frame = +3 Query: 15 EELNADLFRSTLEPVEKALRDAKLDKNAIHEIVLVGGSTRIPKIQKLLQDLFNGKELNKS 194 EEL ADLFRST++PVEK+LRDAK+DK+ +H+IVLVGGSTRIPK+QKLL DLF+GKELNKS Sbjct: 304 EELCADLFRSTMDPVEKSLRDAKMDKSQVHDIVLVGGSTRIPKVQKLLSDLFSGKELNKS 363 Query: 195 INPDEXXXXXXXXXXXILSGDKSEAVQDXXXXXXXXXXXGIETAGGVMTALIKRNTTIPT 374 INPDE ILSGDKSEAVQD GIETAGGVMTALIKRNTTIPT Sbjct: 364 INPDEAVAYGAAVQAAILSGDKSEAVQDLLLLDVAPLSLGIETAGGVMTALIKRNTTIPT 423 Query: 375 KQTQIFSTYSDNQPGVLIQVFEGERAMTKDNNLLGKFELSGIPPAPRGVPQIEVTFDIDA 554 K Q F+TYSDNQPGVLIQV+EGERAMTKDNNLLGKFELSGIPPAPRGVPQIEVTFDIDA Sbjct: 424 KTAQTFTTYSDNQPGVLIQVYEGERAMTKDNNLLGKFELSGIPPAPRGVPQIEVTFDIDA 483 Query: 555 NGILNVSATEKGTGKENKITITNDKGRLSKDDIERMVNXXXXXXXXXXXXXXRIGAKNLL 734 NGILNVSAT+K TGK+NKITITNDKGRLSKDDIERMVN RIGAKN L Sbjct: 484 NGILNVSATDKSTGKQNKITITNDKGRLSKDDIERMVNEAEKYKADDEAQKDRIGAKNGL 543 Query: 735 ESYAYQMKSTIEDEKLKDKISESDRKQIVDRCQEAISWLEKNQQATKEEFDYHYKDLEKL 914 ESYA+ +K TIEDEKLKDKIS D+K+I D+C E + WL+ NQ A KEEF++ KDLE L Sbjct: 544 ESYAFNLKQTIEDEKLKDKISPEDKKKIEDKCDEILKWLDSNQTAEKEEFEHQQKDLEGL 603 Query: 915 CNPIMTKMYQXXXXXXXXXXXXXXXXXXXXXXKRQGPTVEEVD 1043 NPI++K+YQ GPT+EEVD Sbjct: 604 ANPIISKLYQSAGGAPPGAAPGGAAGGAG------GPTIEEVD 640
>sp|P19120|HSP7C_BOVIN Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8) Length = 650 Score = 445 bits (1145), Expect = e-125 Identities = 230/348 (66%), Positives = 262/348 (75%), Gaps = 5/348 (1%) Frame = +3 Query: 15 EELNADLFRSTLEPVEKALRDAKLDKNAIHEIVLVGGSTRIPKIQKLLQDLFNGKELNKS 194 EELNADLFR TL+PVEKALRDAKLDK+ IH+IVLVGGSTRIPKIQKLLQD FNGKELNKS Sbjct: 303 EELNADLFRGTLDPVEKALRDAKLDKSQIHDIVLVGGSTRIPKIQKLLQDFFNGKELNKS 362 Query: 195 INPDEXXXXXXXXXXXILSGDKSEAVQDXXXXXXXXXXXGIETAGGVMTALIKRNTTIPT 374 INPDE ILSGDKSE VQD GIETAGGVMT LIKRNTTIPT Sbjct: 363 INPDEAVAYGAAVQAAILSGDKSENVQDLLLLDVTPLSLGIETAGGVMTVLIKRNTTIPT 422 Query: 375 KQTQIFSTYSDNQPGVLIQVFEGERAMTKDNNLLGKFELSGIPPAPRGVPQIEVTFDIDA 554 KQTQ F+TYSDNQPGVLIQV+EGERAMTKDNNLLGKFEL+GIPPAPRGVPQIEVTFDIDA Sbjct: 423 KQTQTFTTYSDNQPGVLIQVYEGERAMTKDNNLLGKFELTGIPPAPRGVPQIEVTFDIDA 482 Query: 555 NGILNVSATEKGTGKENKITITNDKGRLSKDDIERMVNXXXXXXXXXXXXXXRIGAKNLL 734 NGILNVSA +K TGKENKITITNDKGRLSK+DIERMV ++ +KN L Sbjct: 483 NGILNVSAVDKSTGKENKITITNDKGRLSKEDIERMVQEAEKYKAEDEKQRDKVSSKNSL 542 Query: 735 ESYAYQMKSTIEDEKLKDKISESDRKQIVDRCQEAISWLEKNQQATKEEFDYHYKDLEKL 914 +SYA+ MK+T+EDEKL+ KI++ D+++I+D+C E I+WL+KNQ A KEEF++ K+LEK+ Sbjct: 543 KSYAFNMKATVEDEKLQGKINDEDKQKILDKCNEIINWLDKNQTAEKEEFEHQQKELEKV 602 Query: 915 CNPIMTKMYQ-----XXXXXXXXXXXXXXXXXXXXXXKRQGPTVEEVD 1043 CNPI+TK+YQ GPT+EEVD Sbjct: 603 CNPIITKLYQSAGGMPGGMPGGMPGGFPGGGAPPSGGASSGPTIEEVD 650
>sp|P08108|HSP70_ONCMY Heat shock cognate 70 kDa protein (HSP70) Length = 651 Score = 444 bits (1142), Expect = e-124 Identities = 221/310 (71%), Positives = 252/310 (81%) Frame = +3 Query: 15 EELNADLFRSTLEPVEKALRDAKLDKNAIHEIVLVGGSTRIPKIQKLLQDLFNGKELNKS 194 EELNADLFR TL+PVEK+LRDAK+DK +H+IVLVGGSTRIPKIQKLLQD FNGKELNKS Sbjct: 303 EELNADLFRGTLDPVEKSLRDAKMDKAQVHDIVLVGGSTRIPKIQKLLQDFFNGKELNKS 362 Query: 195 INPDEXXXXXXXXXXXILSGDKSEAVQDXXXXXXXXXXXGIETAGGVMTALIKRNTTIPT 374 INPDE ILSGDKSE VQD GIETAGGVMT LIKRNTTIPT Sbjct: 363 INPDEAVAYGAAVQAAILSGDKSENVQDLLLLDVTPLSLGIETAGGVMTVLIKRNTTIPT 422 Query: 375 KQTQIFSTYSDNQPGVLIQVFEGERAMTKDNNLLGKFELSGIPPAPRGVPQIEVTFDIDA 554 KQTQ F+TYSDNQPGVLIQV+EGERAMTKDNNLLGKFEL+GIPPAPRGVPQIEVTFDIDA Sbjct: 423 KQTQTFTTYSDNQPGVLIQVYEGERAMTKDNNLLGKFELTGIPPAPRGVPQIEVTFDIDA 482 Query: 555 NGILNVSATEKGTGKENKITITNDKGRLSKDDIERMVNXXXXXXXXXXXXXXRIGAKNLL 734 NGI+NVSA +K TGKENKITITNDKGRLSK+DIERMV ++ +KN L Sbjct: 483 NGIMNVSAADKSTGKENKITITNDKGRLSKEDIERMVQEAEKYKCEDDVQRDKVSSKNSL 542 Query: 735 ESYAYQMKSTIEDEKLKDKISESDRKQIVDRCQEAISWLEKNQQATKEEFDYHYKDLEKL 914 ESYA+ MKST+EDEKL+ KIS+ D+ +I+++C E I WL+KNQ A KEE+++H K+LEK+ Sbjct: 543 ESYAFNMKSTVEDEKLQGKISDEDKTKILEKCNEVIGWLDKNQTAEKEEYEHHQKELEKV 602 Query: 915 CNPIMTKMYQ 944 CNPI+TK+YQ Sbjct: 603 CNPIITKLYQ 612
>sp|P08106|HSP70_CHICK Heat shock 70 kDa protein (HSP70) Length = 634 Score = 444 bits (1141), Expect = e-124 Identities = 232/343 (67%), Positives = 257/343 (74%) Frame = +3 Query: 15 EELNADLFRSTLEPVEKALRDAKLDKNAIHEIVLVGGSTRIPKIQKLLQDLFNGKELNKS 194 EELNADLFR TLEPVEKALRDAKLDK I EIVLVGGSTRIPKIQKLLQD FNGKELNKS Sbjct: 306 EELNADLFRGTLEPVEKALRDAKLDKGQIQEIVLVGGSTRIPKIQKLLQDFFNGKELNKS 365 Query: 195 INPDEXXXXXXXXXXXILSGDKSEAVQDXXXXXXXXXXXGIETAGGVMTALIKRNTTIPT 374 INPDE IL GDKSE VQD GIETAGGVMTALIKRNTTIPT Sbjct: 366 INPDEAVAYGAAVQAAILMGDKSENVQDLLLLDVTPLSLGIETAGGVMTALIKRNTTIPT 425 Query: 375 KQTQIFSTYSDNQPGVLIQVFEGERAMTKDNNLLGKFELSGIPPAPRGVPQIEVTFDIDA 554 KQTQ F+TYSDNQ VL+QV+EGERAMTKDNNLLGKF+L+GIPPAPRGVPQIEVTFDIDA Sbjct: 426 KQTQTFTTYSDNQSSVLVQVYEGERAMTKDNNLLGKFDLTGIPPAPRGVPQIEVTFDIDA 485 Query: 555 NGILNVSATEKGTGKENKITITNDKGRLSKDDIERMVNXXXXXXXXXXXXXXRIGAKNLL 734 NGILNVSA +K TGKENKITITNDKGRLSKDDI+RMV R+GAKN L Sbjct: 486 NGILNVSAVDKSTGKENKITITNDKGRLSKDDIDRMVQEAEKYKAEDEANRDRVGAKNSL 545 Query: 735 ESYAYQMKSTIEDEKLKDKISESDRKQIVDRCQEAISWLEKNQQATKEEFDYHYKDLEKL 914 ESY Y MK T+EDEKLK KIS+ D+++++D+CQE IS L++NQ A KEE+++ K+LEKL Sbjct: 546 ESYTYNMKQTVEDEKLKGKISDQDKQKVLDKCQEVISSLDRNQMAEKEEYEHKQKELEKL 605 Query: 915 CNPIMTKMYQXXXXXXXXXXXXXXXXXXXXXXKRQGPTVEEVD 1043 CNPI+TK+YQ GPT+EEVD Sbjct: 606 CNPIVTKLYQGAGGAGAGGSG--------------GPTIEEVD 634
>sp|Q9U639|HSP7D_MANSE Heat shock 70 kDa protein cognate 4 (Hsc 70-4) Length = 652 Score = 443 bits (1139), Expect = e-124 Identities = 224/310 (72%), Positives = 250/310 (80%) Frame = +3 Query: 15 EELNADLFRSTLEPVEKALRDAKLDKNAIHEIVLVGGSTRIPKIQKLLQDLFNGKELNKS 194 EELNADLFRST+EPVEK+LRDAK+DK+ IH+IVLVGGSTRIPK+QKLLQD FNGKELNKS Sbjct: 303 EELNADLFRSTMEPVEKSLRDAKMDKSQIHDIVLVGGSTRIPKVQKLLQDFFNGKELNKS 362 Query: 195 INPDEXXXXXXXXXXXILSGDKSEAVQDXXXXXXXXXXXGIETAGGVMTALIKRNTTIPT 374 INPDE IL GDKSE VQD GIETAGGVMT LIKRNTTIPT Sbjct: 363 INPDEAVAYGAAVQAAILHGDKSEEVQDLLLLDVTPLSLGIETAGGVMTTLIKRNTTIPT 422 Query: 375 KQTQIFSTYSDNQPGVLIQVFEGERAMTKDNNLLGKFELSGIPPAPRGVPQIEVTFDIDA 554 KQTQ F+TYSDNQPGVLIQVFEGERAMTKDNNLLGKFEL+GIPPAPRGVPQIEVTFDIDA Sbjct: 423 KQTQTFTTYSDNQPGVLIQVFEGERAMTKDNNLLGKFELTGIPPAPRGVPQIEVTFDIDA 482 Query: 555 NGILNVSATEKGTGKENKITITNDKGRLSKDDIERMVNXXXXXXXXXXXXXXRIGAKNLL 734 NGILNVSA EK T KENKITITNDKGRLSK++IERMVN I AKN L Sbjct: 483 NGILNVSAVEKSTNKENKITITNDKGRLSKEEIERMVNEAEKYRNEDEKQKETIQAKNAL 542 Query: 735 ESYAYQMKSTIEDEKLKDKISESDRKQIVDRCQEAISWLEKNQQATKEEFDYHYKDLEKL 914 ESY + MKST+EDEKLKDKIS+SD++ I+D+C + I WL+ NQ A KEE+++ K+LE + Sbjct: 543 ESYCFNMKSTMEDEKLKDKISDSDKQTILDKCNDTIKWLDSNQLADKEEYEHKQKELEGI 602 Query: 915 CNPIMTKMYQ 944 CNPI+TK+YQ Sbjct: 603 CNPIITKLYQ 612
>sp|Q24789|HSP70_ECHGR Heat shock cognate 70 kDa protein (HSP70) Length = 665 Score = 442 bits (1137), Expect = e-124 Identities = 220/310 (70%), Positives = 253/310 (81%) Frame = +3 Query: 15 EELNADLFRSTLEPVEKALRDAKLDKNAIHEIVLVGGSTRIPKIQKLLQDLFNGKELNKS 194 EEL +DLFRSTL+PVEKALRDAKLDK A+HEIVLVGGSTRIPK+QKLLQD FNG+ELNKS Sbjct: 305 EELCSDLFRSTLDPVEKALRDAKLDKGAVHEIVLVGGSTRIPKVQKLLQDFFNGRELNKS 364 Query: 195 INPDEXXXXXXXXXXXILSGDKSEAVQDXXXXXXXXXXXGIETAGGVMTALIKRNTTIPT 374 INPDE IL+GDKSEAVQD G+ETAGGVMTALIKRNTTIPT Sbjct: 365 INPDEAVAYGAAVQAAILTGDKSEAVQDLLLLDVAPLSLGLETAGGVMTALIKRNTTIPT 424 Query: 375 KQTQIFSTYSDNQPGVLIQVFEGERAMTKDNNLLGKFELSGIPPAPRGVPQIEVTFDIDA 554 KQTQ F+TYSDNQPGVLIQV+EGERAM +DNNLLGKFELSGIPP PRGVPQIEVTFDIDA Sbjct: 425 KQTQTFTTYSDNQPGVLIQVYEGERAMKRDNNLLGKFELSGIPPGPRGVPQIEVTFDIDA 484 Query: 555 NGILNVSATEKGTGKENKITITNDKGRLSKDDIERMVNXXXXXXXXXXXXXXRIGAKNLL 734 NGILNVSA +K TGK+NKITIT DKGRLSK++IERMVN R+ AKN L Sbjct: 485 NGILNVSAVDKSTGKQNKITITRDKGRLSKEEIERMVNDAEKFKQEDEKQRDRVAAKNGL 544 Query: 735 ESYAYQMKSTIEDEKLKDKISESDRKQIVDRCQEAISWLEKNQQATKEEFDYHYKDLEKL 914 ESYA+ MKST+EDEK+K+KI ESDR++I+++C+E + WL+ NQQA KEE+++ K+LE + Sbjct: 545 ESYAFSMKSTVEDEKVKEKIGESDRRRIMEKCEETVKWLDGNQQAEKEEYEHRQKELESV 604 Query: 915 CNPIMTKMYQ 944 CNPI+ KMYQ Sbjct: 605 CNPIIAKMYQ 614
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 108,351,940
Number of Sequences: 369166
Number of extensions: 2050111
Number of successful extensions: 7572
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6586
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6883
length of database: 68,354,980
effective HSP length: 112
effective length of database: 47,664,660
effective search space used: 12011494320
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail