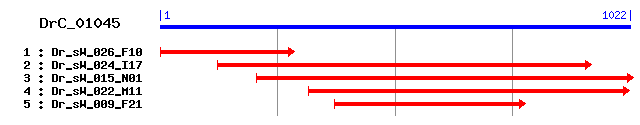
DrC_01045
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01045 (1022 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q94547|RGA_DROME Regulator of gene activity (Regena prot... 82 3e-15 sp|Q9NZN8|CNOT2_HUMAN CCR4-NOT transcription complex subuni... 75 3e-13 sp|Q8C5L3|CNOT2_MOUSE CCR4-NOT transcription complex subuni... 75 3e-13 sp|O75175|CNOT3_HUMAN CCR4-NOT transcription complex subuni... 63 2e-09 sp|Q8K0V4|CNOT3_MOUSE CCR4-NOT transcription complex subuni... 63 2e-09 sp|P06100|NOT2_YEAST General negative regulator of transcri... 55 2e-07 sp|Q12514|NOT5_YEAST General negative regulator of transcri... 40 0.014 sp|P04936|POLG_HRV2 Genome polyprotein [Contains: Coat prot... 35 0.26 sp|O43918|AIRE_HUMAN Autoimmune regulator (Autoimmune polye... 34 0.59 sp|P36304|POLR_KYMVJ RNA replicase polyprotein 32 2.9
>sp|Q94547|RGA_DROME Regulator of gene activity (Regena protein) Length = 585 Score = 81.6 bits (200), Expect = 3e-15 Identities = 38/109 (34%), Positives = 68/109 (62%) Frame = +2 Query: 20 RPHDMDYAVPKEYRIRHSILDKLADPLPEKLSDETLFWIFYNLTGEECQTTAAKQLMARQ 199 R D+++ VP EY I +I DKL P+ +KL ++ LF++FY G+ Q AA +L +R+ Sbjct: 458 RAQDVEFNVPPEYLINFAIRDKLTAPVLKKLQEDLLFFLFYTNIGDMMQLMAAAELHSRE 517 Query: 200 WRYNKRDKFWLTRDSKGVNEPTVHPNLERGSYIVWDYEKAQRVTKILTV 346 WRY+ +K W+TR G+++ + ERG++ +D + +R++K+ + Sbjct: 518 WRYHVEEKIWITR-IPGIDQYEKNGTKERGTFYYFDAQSWKRLSKVFQI 565
>sp|Q9NZN8|CNOT2_HUMAN CCR4-NOT transcription complex subunit 2 (CCR4-associated factor 2) Length = 540 Score = 75.1 bits (183), Expect = 3e-13 Identities = 44/116 (37%), Positives = 66/116 (56%), Gaps = 1/116 (0%) Frame = +2 Query: 20 RPHDMDYAVPKEYRIRHSILDKLADPLPEKLSDETLFWIFYNLTGEECQTTAAKQLMARQ 199 RP D+D+ VP EY I DKLA + ++ LF+++Y G+ Q AA +L R Sbjct: 409 RPQDIDFHVPSEYLTNIHIRDKLAAIKLGRYGEDLLFYLYYMNGGDVLQLLAAVELFNRD 468 Query: 200 WRYNKRDKFWLTRDSKGVNEPTVHPN-LERGSYIVWDYEKAQRVTKILTVYYKDLE 364 WRY+K ++ W+TR + G+ EPT+ N ERG+Y +D ++V K + Y LE Sbjct: 469 WRYHKEERVWITR-APGM-EPTMKTNTYERGTYYFFDCLNWRKVAKEFHLEYDKLE 522
>sp|Q8C5L3|CNOT2_MOUSE CCR4-NOT transcription complex subunit 2 (CCR4-associated factor 2) Length = 540 Score = 75.1 bits (183), Expect = 3e-13 Identities = 44/116 (37%), Positives = 66/116 (56%), Gaps = 1/116 (0%) Frame = +2 Query: 20 RPHDMDYAVPKEYRIRHSILDKLADPLPEKLSDETLFWIFYNLTGEECQTTAAKQLMARQ 199 RP D+D+ VP EY I DKLA + ++ LF+++Y G+ Q AA +L R Sbjct: 409 RPQDIDFHVPSEYLTNIHIRDKLAAIKLGRYGEDLLFYLYYMNGGDVLQLLAAVELFNRD 468 Query: 200 WRYNKRDKFWLTRDSKGVNEPTVHPN-LERGSYIVWDYEKAQRVTKILTVYYKDLE 364 WRY+K ++ W+TR + G+ EPT+ N ERG+Y +D ++V K + Y LE Sbjct: 469 WRYHKEERVWITR-APGM-EPTMKTNTYERGTYYFFDCLNWRKVAKEFHLEYDKLE 522
>sp|O75175|CNOT3_HUMAN CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3) Length = 753 Score = 62.8 bits (151), Expect = 2e-09 Identities = 35/89 (39%), Positives = 51/89 (57%), Gaps = 1/89 (1%) Frame = +2 Query: 104 EKLSDETLFWIFYNLTGEECQTTAAKQLMARQWRYNKRDKFWLTRDSKGVNEPTVHPNLE 283 ++LS ETLF+IFY L G + Q AAK L + WR++ + W R + T+ E Sbjct: 664 QRLSTETLFFIFYYLEGTKAQYLAAKALKKQSWRFHTKYMMWFQRHEE---PKTITDEFE 720 Query: 284 RGSYIVWDYEK-AQRVTKILTVYYKDLEN 367 +G+YI +DYEK QR + T Y+ LE+ Sbjct: 721 QGTYIYFDYEKWGQRKKEGFTFEYRYLED 749
>sp|Q8K0V4|CNOT3_MOUSE CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3) Length = 751 Score = 62.8 bits (151), Expect = 2e-09 Identities = 35/89 (39%), Positives = 51/89 (57%), Gaps = 1/89 (1%) Frame = +2 Query: 104 EKLSDETLFWIFYNLTGEECQTTAAKQLMARQWRYNKRDKFWLTRDSKGVNEPTVHPNLE 283 ++LS ETLF+IFY L G + Q AAK L + WR++ + W R + T+ E Sbjct: 662 QRLSTETLFFIFYYLEGTKAQYLAAKALKKQSWRFHTKYMMWFQRHEE---PKTITDEFE 718 Query: 284 RGSYIVWDYEK-AQRVTKILTVYYKDLEN 367 +G+YI +DYEK QR + T Y+ LE+ Sbjct: 719 QGTYIYFDYEKWGQRKKEGFTFEYRYLED 747
>sp|P06100|NOT2_YEAST General negative regulator of transcription subunit 2 Length = 191 Score = 55.5 bits (132), Expect = 2e-07 Identities = 30/81 (37%), Positives = 46/81 (56%), Gaps = 2/81 (2%) Frame = +2 Query: 116 DETLFWIFYNLTGEECQTTAAKQLMARQWRYNKRDKFWLTRDSKGVNEPTVHPN--LERG 289 DETLF++FY G Q +L R WRY+K K WLT+D + EP V + ERG Sbjct: 109 DETLFFLFYKHPGTVIQELTYLELRKRNWRYHKTLKAWLTKDP--MMEPIVSADGLSERG 166 Query: 290 SYIVWDYEKAQRVTKILTVYY 352 SY+ +D ++ ++ + ++Y Sbjct: 167 SYVFFDPQRWEKCQRDFLLFY 187
>sp|Q12514|NOT5_YEAST General negative regulator of transcription subunit 5 Length = 560 Score = 39.7 bits (91), Expect = 0.014 Identities = 24/72 (33%), Positives = 39/72 (54%), Gaps = 1/72 (1%) Frame = +2 Query: 107 KLSDETLFWIFYNLTGEECQTTAAKQLMA-RQWRYNKRDKFWLTRDSKGVNEPTVHPNLE 283 K +TLF+IFY+ G Q AA++L R W +NK D+ W ++ + + P E Sbjct: 471 KFDLDTLFFIFYHYQGSYEQFLAARELFKNRNWLFNKVDRCWYYKEIEKL--PPGMGKSE 528 Query: 284 RGSYIVWDYEKA 319 S+ +DY+K+ Sbjct: 529 EESWRYFDYKKS 540
>sp|P04936|POLG_HRV2 Genome polyprotein [Contains: Coat protein VP4 (P1A); Coat protein VP2 (P1B); Coat protein VP3 (P1C); Coat protein VP1 (P1D); Picornain 2A (Core protein P2A); Core protein P2B; Core protein P2C; Core protein P3A; Genome-linked protein VPg (P3B); Picornain 3C (Protease 3C) (P3C); RNA-directed RNA polymerase (P3D)] Length = 2150 Score = 35.4 bits (80), Expect = 0.26 Identities = 21/93 (22%), Positives = 38/93 (40%), Gaps = 1/93 (1%) Frame = +2 Query: 209 NKRDKFWLTRDSKGVNEPTVHPNLERGSYIVWDYEKAQRVTKILT-VYYKDLENVTPATT 385 N K L G+ EPT + G++++WD ++ ++ + N +P + Sbjct: 454 NTTVKLLLAYTPPGIAEPTTRKDAMLGTHVIWDVGLQSTISMVVPWISASHYRNTSPGRS 513 Query: 386 KSLFPFCYNSPFIKRSAVNSSAIPPPQLPPTGR 484 S + C+ + + PPQ PPT R Sbjct: 514 TSGYITCW---------YQTRLVIPPQTPPTAR 537
>sp|O43918|AIRE_HUMAN Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein) Length = 545 Score = 34.3 bits (77), Expect = 0.59 Identities = 47/200 (23%), Positives = 86/200 (43%) Frame = +2 Query: 41 AVPKEYRIRHSILDKLADPLPEKLSDETLFWIFYNLTGEECQTTAAKQLMARQWRYNKRD 220 AV + + H++ D D +PE ETL +L +E A L++ Sbjct: 21 AVDSAFPLLHALADH--DVVPEDKFQETL-----HLKEKEGCPQAFHALLS--------- 64 Query: 221 KFWLTRDSKGVNEPTVHPNLERGSYIVWDYEKAQRVTKILTVYYKDLENVTPATTKSLFP 400 + LT+DS + + R + ++ E+ R+ IL + KD++ P + Sbjct: 65 -WLLTQDSTAILD------FWRVLFKDYNLERYGRLQPILDSFPKDVDLSQPRKGR---- 113 Query: 401 FCYNSPFIKRSAVNSSAIPPPQLPPTGRTNQKNLATFLQNPSAAASVEARLLNNPISQNF 580 K AV + +PPP+LP + +++ A +A A++ R +P SQ Sbjct: 114 --------KPPAVPKALVPPPRLPTKRKASEEARA------AAPAALTPRGTASPGSQLK 159 Query: 581 SQPDQMMTSTASQIRVQINN 640 ++P + S+A Q R+ + N Sbjct: 160 AKPPKKPESSAEQQRLPLGN 179
>sp|P36304|POLR_KYMVJ RNA replicase polyprotein Length = 1874 Score = 32.0 bits (71), Expect = 2.9 Identities = 23/75 (30%), Positives = 35/75 (46%), Gaps = 6/75 (8%) Frame = +2 Query: 410 NSPFIKRSAVNSSAIPPPQLPPTGRTNQKNLATFLQNPSAAASVEARLLNNP------IS 571 +SPF+ +N SA+PPPQ PT T + L P V++ L+ +P +S Sbjct: 722 HSPFLSDGQLNYSALPPPQ-DPTNTT-----LSLLPEPKPPTEVQSPLMADPTCVGPAVS 775 Query: 572 QNFSQPDQMMTSTAS 616 + P +TAS Sbjct: 776 FSSLYPRDFFPNTAS 790
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 107,919,087
Number of Sequences: 369166
Number of extensions: 2144907
Number of successful extensions: 5640
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5382
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5631
length of database: 68,354,980
effective HSP length: 111
effective length of database: 47,849,395
effective search space used: 10957511455
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail