DrC_01014
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01014 (1049 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O02649|CH60_DROME 60 kDa heat shock protein, mitochondri... 538 e-153 sp|P10809|CH60_HUMAN 60 kDa heat shock protein, mitochondri... 531 e-150 sp|P18687|CH60_CRIGR 60 kDa heat shock protein, mitochondri... 531 e-150 sp|P63039|CH60_RAT 60 kDa heat shock protein, mitochondrial... 530 e-150 sp|Q5NVM5|CH60_PONPY 60 kDa heat shock protein, mitochondri... 529 e-150 sp|Q9VMN5|CH60C_DROME Probable 60 kDa heat shock protein ho... 516 e-146 sp|P50140|CH60_CAEEL Chaperonin homolog HSP60, mitochondria... 498 e-140 sp|P25420|CH63_HELVI 63 kDa chaperonin, mitochondrial precu... 455 e-127 sp|O74261|HSP60_CANAL Heat shock protein 60, mitochondrial ... 414 e-115 sp|Q9VPS5|CH60B_DROME Probable 60 kDa heat shock protein ho... 412 e-115
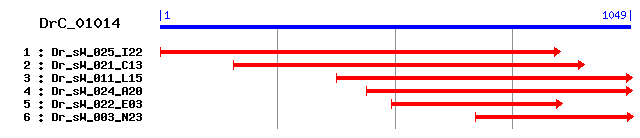
>sp|O02649|CH60_DROME 60 kDa heat shock protein, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (CPN60) (Heat shock protein 60) (HSP-60) (Mitochondrial matrix protein P1) Length = 573 Score = 538 bits (1387), Expect = e-153 Identities = 267/349 (76%), Positives = 316/349 (90%) Frame = +2 Query: 2 RSAMLVGVDILADAVAVTMGPKGRNVILEKSWGSPKITKDGVTVAKDIELKDKFQNIGAK 181 R+ ML GVD+LADAVAVTMGPKGRNVI+E+SWGSPKITKDGVTVAK IELKDKFQNIGAK Sbjct: 33 RAMMLQGVDVLADAVAVTMGPKGRNVIIEQSWGSPKITKDGVTVAKSIELKDKFQNIGAK 92 Query: 182 LVQDVANNTNEEAGDGTTAATVLARAIAKEGFEKISKGANPIDFRRGVMMAVDNVVASLK 361 LVQDVANNTNEEAGDGTT ATVLARAIAKEGFEKISKGANP++ RRGVM+AV+ V +LK Sbjct: 93 LVQDVANNTNEEAGDGTTTATVLARAIAKEGFEKISKGANPVEIRRGVMLAVETVKDNLK 152 Query: 362 AMSKPVTTPEEIAQVATISANGDRSIGDLISEAMKKVGKDGTITVKDGKTLHDELEVIEG 541 MS+PV+TPEEIAQVATISANGD++IG+LISEAMKKVG+DG ITVKDGKTL DELEVIEG Sbjct: 153 TMSRPVSTPEEIAQVATISANGDQAIGNLISEAMKKVGRDGVITVKDGKTLTDELEVIEG 212 Query: 542 MKFDRGYISPYFINTDKGAKCEFQDAFILFSEKKISSIQSLIPVLEMTHQQKKPLLIVAE 721 MKFDRGYISPYFIN+ KGAK EFQDA +L SEKKISS+QS+IP LE+ + Q+KPL+I+AE Sbjct: 213 MKFDRGYISPYFINSSKGAKVEFQDALLLLSEKKISSVQSIIPALELANAQRKPLVIIAE 272 Query: 722 DVDGEALTALVLNRLKLGLQVCAVKAPGFGDNRKNTLRDMATSTGGIVFGDEADTYKIEN 901 D+DGEAL+ LV+NRLK+GLQV AVKAPGFGDNRK+TL DMA ++GGIVFGD+AD K+E+ Sbjct: 273 DIDGEALSTLVVNRLKIGLQVAAVKAPGFGDNRKSTLTDMAIASGGIVFGDDADLVKLED 332 Query: 902 VQLHDLGRAQEILITKDDTLVMKGKGLQKDIQKRIDEIRDEIALSTSEY 1048 V++ DLG+ E++ITKDDTL++KGKG + D+ +R ++I+D+I +TSEY Sbjct: 333 VKVSDLGQVGEVVITKDDTLLLKGKGKKDDVLRRANQIKDQIEDTTSEY 381
>sp|P10809|CH60_HUMAN 60 kDa heat shock protein, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (CPN60) (Heat shock protein 60) (HSP-60) (Mitochondrial matrix protein P1) (P60 lymphocyte protein) (HuCHA60) Length = 573 Score = 531 bits (1368), Expect = e-150 Identities = 260/349 (74%), Positives = 310/349 (88%) Frame = +2 Query: 2 RSAMLVGVDILADAVAVTMGPKGRNVILEKSWGSPKITKDGVTVAKDIELKDKFQNIGAK 181 R+ ML GVD+LADAVAVTMGPKGR VI+E+SWGSPK+TKDGVTVAK I+LKDK++NIGAK Sbjct: 37 RALMLQGVDLLADAVAVTMGPKGRTVIIEQSWGSPKVTKDGVTVAKSIDLKDKYKNIGAK 96 Query: 182 LVQDVANNTNEEAGDGTTAATVLARAIAKEGFEKISKGANPIDFRRGVMMAVDNVVASLK 361 LVQDVANNTNEEAGDGTT ATVLAR+IAKEGFEKISKGANP++ RRGVM+AVD V+A LK Sbjct: 97 LVQDVANNTNEEAGDGTTTATVLARSIAKEGFEKISKGANPVEIRRGVMLAVDAVIAELK 156 Query: 362 AMSKPVTTPEEIAQVATISANGDRSIGDLISEAMKKVGKDGTITVKDGKTLHDELEVIEG 541 SKPVTTPEEIAQVATISANGD+ IG++IS+AMKKVG+ G ITVKDGKTL+DELE+IEG Sbjct: 157 KQSKPVTTPEEIAQVATISANGDKEIGNIISDAMKKVGRKGVITVKDGKTLNDELEIIEG 216 Query: 542 MKFDRGYISPYFINTDKGAKCEFQDAFILFSEKKISSIQSLIPVLEMTHQQKKPLLIVAE 721 MKFDRGYISPYFINT KG KCEFQDA++L SEKKISSIQS++P LE+ + +KPL+I+AE Sbjct: 217 MKFDRGYISPYFINTSKGQKCEFQDAYVLLSEKKISSIQSIVPALEIANAHRKPLVIIAE 276 Query: 722 DVDGEALTALVLNRLKLGLQVCAVKAPGFGDNRKNTLRDMATSTGGIVFGDEADTYKIEN 901 DVDGEAL+ LVLNRLK+GLQV AVKAPGFGDNRKN L+DMA +TGG VFG+E T +E+ Sbjct: 277 DVDGEALSTLVLNRLKVGLQVVAVKAPGFGDNRKNQLKDMAIATGGAVFGEEGLTLNLED 336 Query: 902 VQLHDLGRAQEILITKDDTLVMKGKGLQKDIQKRIDEIRDEIALSTSEY 1048 VQ HDLG+ E+++TKDD +++KGKG + I+KRI EI +++ ++TSEY Sbjct: 337 VQPHDLGKVGEVIVTKDDAMLLKGKGDKAQIEKRIQEIIEQLDVTTSEY 385
>sp|P18687|CH60_CRIGR 60 kDa heat shock protein, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (CPN60) (Heat shock protein 60) (HSP-60) (Mitochondrial matrix protein P1) Length = 573 Score = 531 bits (1367), Expect = e-150 Identities = 258/349 (73%), Positives = 309/349 (88%) Frame = +2 Query: 2 RSAMLVGVDILADAVAVTMGPKGRNVILEKSWGSPKITKDGVTVAKDIELKDKFQNIGAK 181 R+ ML GVD+LADAVAVTMGPKGR VI+E+SWGSPK+TKDGVTVAK I+LKDK++NIGAK Sbjct: 37 RALMLQGVDLLADAVAVTMGPKGRTVIIEQSWGSPKVTKDGVTVAKAIDLKDKYKNIGAK 96 Query: 182 LVQDVANNTNEEAGDGTTAATVLARAIAKEGFEKISKGANPIDFRRGVMMAVDNVVASLK 361 LVQDVANNTNEEAGDGTT ATVLAR+IAKEGFEKISKGANP++ RRGVM+AVD V+A LK Sbjct: 97 LVQDVANNTNEEAGDGTTTATVLARSIAKEGFEKISKGANPVEIRRGVMLAVDAVIAELK 156 Query: 362 AMSKPVTTPEEIAQVATISANGDRSIGDLISEAMKKVGKDGTITVKDGKTLHDELEVIEG 541 SKPVTTPEEIAQVATISANGD+ IG++IS+AMKKVG+ G ITVKDGKTL+DELE+IEG Sbjct: 157 KQSKPVTTPEEIAQVATISANGDKDIGNIISDAMKKVGRKGVITVKDGKTLNDELEIIEG 216 Query: 542 MKFDRGYISPYFINTDKGAKCEFQDAFILFSEKKISSIQSLIPVLEMTHQQKKPLLIVAE 721 MKFDRGYISPYFINT KG KCEFQDA++L SEKKISS+QS++P LE+ + +KPL+I+AE Sbjct: 217 MKFDRGYISPYFINTSKGQKCEFQDAYVLLSEKKISSVQSIVPALEIANAHRKPLVIIAE 276 Query: 722 DVDGEALTALVLNRLKLGLQVCAVKAPGFGDNRKNTLRDMATSTGGIVFGDEADTYKIEN 901 DVDGEAL+ LVLNRLK+GLQV AVKAPGFGDNRKN L+DMA +TGG VFG+E +E+ Sbjct: 277 DVDGEALSTLVLNRLKVGLQVVAVKAPGFGDNRKNQLKDMAIATGGAVFGEEGLNLNLED 336 Query: 902 VQLHDLGRAQEILITKDDTLVMKGKGLQKDIQKRIDEIRDEIALSTSEY 1048 VQ HDLG+ E+++TKDD +++KGKG + I+KRI EI +++ ++TSEY Sbjct: 337 VQAHDLGKVGEVIVTKDDAMLLKGKGEKAQIEKRIQEITEQLEITTSEY 385
>sp|P63039|CH60_RAT 60 kDa heat shock protein, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (CPN60) (Heat shock protein 60) (HSP-60) (Mitochondrial matrix protein P1) (HSP-65) sp|P63038|CH60_MOUSE 60 kDa heat shock protein, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (CPN60) (Heat shock protein 60) (HSP-60) (Mitochondrial matrix protein P1) (HSP-65) Length = 573 Score = 530 bits (1366), Expect = e-150 Identities = 258/349 (73%), Positives = 309/349 (88%) Frame = +2 Query: 2 RSAMLVGVDILADAVAVTMGPKGRNVILEKSWGSPKITKDGVTVAKDIELKDKFQNIGAK 181 R+ ML GVD+LADAVAVTMGPKGR VI+E+SWGSPK+TKDGVTVAK I+LKDK++NIGAK Sbjct: 37 RALMLQGVDLLADAVAVTMGPKGRTVIIEQSWGSPKVTKDGVTVAKSIDLKDKYKNIGAK 96 Query: 182 LVQDVANNTNEEAGDGTTAATVLARAIAKEGFEKISKGANPIDFRRGVMMAVDNVVASLK 361 LVQDVANNTNEEAGDGTT ATVLAR+IAKEGFEKISKGANP++ RRGVM+AVD V+A LK Sbjct: 97 LVQDVANNTNEEAGDGTTTATVLARSIAKEGFEKISKGANPVEIRRGVMLAVDAVIAELK 156 Query: 362 AMSKPVTTPEEIAQVATISANGDRSIGDLISEAMKKVGKDGTITVKDGKTLHDELEVIEG 541 SKPVTTPEEIAQVATISANGD+ IG++IS+AMKKVG+ G ITVKDGKTL+DELE+IEG Sbjct: 157 KQSKPVTTPEEIAQVATISANGDKDIGNIISDAMKKVGRKGVITVKDGKTLNDELEIIEG 216 Query: 542 MKFDRGYISPYFINTDKGAKCEFQDAFILFSEKKISSIQSLIPVLEMTHQQKKPLLIVAE 721 MKFDRGYISPYFINT KG KCEFQDA++L SEKKISS+QS++P LE+ + +KPL+I+AE Sbjct: 217 MKFDRGYISPYFINTSKGQKCEFQDAYVLLSEKKISSVQSIVPALEIANAHRKPLVIIAE 276 Query: 722 DVDGEALTALVLNRLKLGLQVCAVKAPGFGDNRKNTLRDMATSTGGIVFGDEADTYKIEN 901 DVDGEAL+ LVLNRLK+GLQV AVKAPGFGDNRKN L+DMA +TGG VFG+E +E+ Sbjct: 277 DVDGEALSTLVLNRLKVGLQVVAVKAPGFGDNRKNQLKDMAIATGGAVFGEEGLNLNLED 336 Query: 902 VQLHDLGRAQEILITKDDTLVMKGKGLQKDIQKRIDEIRDEIALSTSEY 1048 VQ HDLG+ E+++TKDD +++KGKG + I+KRI EI +++ ++TSEY Sbjct: 337 VQAHDLGKVGEVIVTKDDAMLLKGKGDKAHIEKRIQEITEQLDITTSEY 385
>sp|Q5NVM5|CH60_PONPY 60 kDa heat shock protein, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (CPN60) (Heat shock protein 60) (HSP-60) Length = 573 Score = 529 bits (1362), Expect = e-150 Identities = 258/349 (73%), Positives = 310/349 (88%) Frame = +2 Query: 2 RSAMLVGVDILADAVAVTMGPKGRNVILEKSWGSPKITKDGVTVAKDIELKDKFQNIGAK 181 R+ ML GVD+LADAVAVTMGPKGR VI+E+SWGSPK+TKDGVTVAK I+LKDK++NIGAK Sbjct: 37 RALMLRGVDLLADAVAVTMGPKGRTVIIEQSWGSPKVTKDGVTVAKSIDLKDKYKNIGAK 96 Query: 182 LVQDVANNTNEEAGDGTTAATVLARAIAKEGFEKISKGANPIDFRRGVMMAVDNVVASLK 361 LVQDVANNTNEEAGDGTT ATVLAR+IAKEGFEKISKGANP++ RRGVM+AVD V+A LK Sbjct: 97 LVQDVANNTNEEAGDGTTTATVLARSIAKEGFEKISKGANPVEIRRGVMLAVDAVIAELK 156 Query: 362 AMSKPVTTPEEIAQVATISANGDRSIGDLISEAMKKVGKDGTITVKDGKTLHDELEVIEG 541 SKPVTTPEEIAQVATISANGD+ IG++IS+AMKKVG+ G ITVKDGKTL+DELE+IEG Sbjct: 157 KQSKPVTTPEEIAQVATISANGDKEIGNIISDAMKKVGRKGVITVKDGKTLNDELEIIEG 216 Query: 542 MKFDRGYISPYFINTDKGAKCEFQDAFILFSEKKISSIQSLIPVLEMTHQQKKPLLIVAE 721 MKFDRGYISPYFINT KG KCEFQDA++L SEKKISS+QS++P LE+ + +KPL+I+AE Sbjct: 217 MKFDRGYISPYFINTSKGQKCEFQDAYVLLSEKKISSVQSIVPALEIANAHRKPLVIIAE 276 Query: 722 DVDGEALTALVLNRLKLGLQVCAVKAPGFGDNRKNTLRDMATSTGGIVFGDEADTYKIEN 901 DVDGEAL+ LVLNRLK+GLQV AVKAPGFGDNRKN L+DMA +TGG VFG+E T +E+ Sbjct: 277 DVDGEALSTLVLNRLKVGLQVVAVKAPGFGDNRKNQLKDMAIATGGAVFGEEGLTLNLED 336 Query: 902 VQLHDLGRAQEILITKDDTLVMKGKGLQKDIQKRIDEIRDEIALSTSEY 1048 VQ HDLG+ E+++TKDD ++++GKG + I+KRI EI +++ ++TSEY Sbjct: 337 VQPHDLGKVGEVIVTKDDAMLLEGKGDKAQIEKRIQEIIEQLDVTTSEY 385
>sp|Q9VMN5|CH60C_DROME Probable 60 kDa heat shock protein homolog 2, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (CPN60) (Heat shock protein 60) (HSP-60) Length = 576 Score = 516 bits (1328), Expect = e-146 Identities = 250/349 (71%), Positives = 306/349 (87%) Frame = +2 Query: 2 RSAMLVGVDILADAVAVTMGPKGRNVILEKSWGSPKITKDGVTVAKDIELKDKFQNIGAK 181 R+ ML GVD+LADAVAVTMGPKGRNVI+E+SWGSPKITKDGVTVAK I LKDKFQNIGAK Sbjct: 37 RAMMLQGVDVLADAVAVTMGPKGRNVIIEQSWGSPKITKDGVTVAKSIALKDKFQNIGAK 96 Query: 182 LVQDVANNTNEEAGDGTTAATVLARAIAKEGFEKISKGANPIDFRRGVMMAVDNVVASLK 361 LVQDVANNTNEEAGDGTT ATVLARAIAKEGFEKIS+GA+P++ RRGVM+A++ V +L+ Sbjct: 97 LVQDVANNTNEEAGDGTTTATVLARAIAKEGFEKISRGASPVEIRRGVMLAIETVKDNLR 156 Query: 362 AMSKPVTTPEEIAQVATISANGDRSIGDLISEAMKKVGKDGTITVKDGKTLHDELEVIEG 541 +S+PV TPEEI QVATISANGD+S+G+LISEA+KKVG+DG ITVKDGKTL DELEVIEG Sbjct: 157 RLSRPVNTPEEICQVATISANGDKSVGNLISEAIKKVGRDGVITVKDGKTLCDELEVIEG 216 Query: 542 MKFDRGYISPYFINTDKGAKCEFQDAFILFSEKKISSIQSLIPVLEMTHQQKKPLLIVAE 721 MKFDRGYISPYFINT KGAK EFQDA +LF EKKI S S++P LE+ + Q+KPL+I+AE Sbjct: 217 MKFDRGYISPYFINTSKGAKVEFQDALLLFCEKKIKSAPSIVPALELANAQRKPLVIIAE 276 Query: 722 DVDGEALTALVLNRLKLGLQVCAVKAPGFGDNRKNTLRDMATSTGGIVFGDEADTYKIEN 901 D++ EAL+ LV+NRLK+GLQVCAVKAPGFGDNRK L DMA +TGGIVFGDEA+ ++E+ Sbjct: 277 DLEAEALSTLVVNRLKVGLQVCAVKAPGFGDNRKENLMDMAVATGGIVFGDEANMVRLED 336 Query: 902 VQLHDLGRAQEILITKDDTLVMKGKGLQKDIQKRIDEIRDEIALSTSEY 1048 +++ D GR E++++KDDT+++KGKG + +++KR++ +R+ I STS Y Sbjct: 337 IKMSDFGRVGEVVVSKDDTMLLKGKGQKAEVEKRVEGLREAIKESTSSY 385
>sp|P50140|CH60_CAEEL Chaperonin homolog HSP60, mitochondrial precursor (Heat shock protein 60) (HSP-60) Length = 568 Score = 498 bits (1281), Expect = e-140 Identities = 245/349 (70%), Positives = 298/349 (85%) Frame = +2 Query: 2 RSAMLVGVDILADAVAVTMGPKGRNVILEKSWGSPKITKDGVTVAKDIELKDKFQNIGAK 181 R AMLVGV++LADAV+VTMGPKGRNVI+E+SWGSPKITKDGVTVAK I+LKDK+QN+GAK Sbjct: 28 RQAMLVGVNLLADAVSVTMGPKGRNVIIEQSWGSPKITKDGVTVAKSIDLKDKYQNLGAK 87 Query: 182 LVQDVANNTNEEAGDGTTAATVLARAIAKEGFEKISKGANPIDFRRGVMMAVDNVVASLK 361 L+QDVAN NEEAGDGTT ATVLARAIAKEGFE I +G N ++ RRGVM AV+ VVA LK Sbjct: 88 LIQDVANKANEEAGDGTTCATVLARAIAKEGFESIRQGGNAVEIRRGVMNAVEVVVAELK 147 Query: 362 AMSKPVTTPEEIAQVATISANGDRSIGDLISEAMKKVGKDGTITVKDGKTLHDELEVIEG 541 +SK VTTPEEIAQVATISANGD +G+LIS+AMKKVG G ITVKDGKTL+DELE+IEG Sbjct: 148 KISKKVTTPEEIAQVATISANGDTVVGNLISDAMKKVGTTGVITVKDGKTLNDELELIEG 207 Query: 542 MKFDRGYISPYFINTDKGAKCEFQDAFILFSEKKISSIQSLIPVLEMTHQQKKPLLIVAE 721 MKFDRGYISPYFI + KGAK E++ A +L SEKKIS +Q ++P LE+ ++ ++PL+I+AE Sbjct: 208 MKFDRGYISPYFITSAKGAKVEYEKALVLLSEKKISQVQDIVPALELANKLRRPLVIIAE 267 Query: 722 DVDGEALTALVLNRLKLGLQVCAVKAPGFGDNRKNTLRDMATSTGGIVFGDEADTYKIEN 901 DVDGEALT LVLNRLK+GLQV A+KAPGFGDNRKNTL+DM +TG +FGD+++ KIE+ Sbjct: 268 DVDGEALTTLVLNRLKVGLQVVAIKAPGFGDNRKNTLKDMGIATGATIFGDDSNLIKIED 327 Query: 902 VQLHDLGRAQEILITKDDTLVMKGKGLQKDIQKRIDEIRDEIALSTSEY 1048 + +DLG E+ ITKDDTL+++G+G Q +I+KRI+ I DEI STS+Y Sbjct: 328 ITANDLGEVDEVTITKDDTLLLRGRGDQTEIEKRIEHITDEIEQSTSDY 376
>sp|P25420|CH63_HELVI 63 kDa chaperonin, mitochondrial precursor (p63) Length = 569 Score = 455 bits (1170), Expect = e-127 Identities = 231/349 (66%), Positives = 281/349 (80%) Frame = +2 Query: 2 RSAMLVGVDILADAVAVTMGPKGRNVILEKSWGSPKITKDGVTVAKDIELKDKFQNIGAK 181 RS ML GVDILADA VTMGPKG NVIL K+ G PKITKDGVTVAK I+LKDKFQNIGA+ Sbjct: 40 RSLMLQGVDILADADDVTMGPKGVNVILAKNLGPPKITKDGVTVAKGIDLKDKFQNIGAR 99 Query: 182 LVQDVANNTNEEAGDGTTAATVLARAIAKEGFEKISKGANPIDFRRGVMMAVDNVVASLK 361 LVQ+VAN TNEEAGDGTT ATVLAR IAKEGFE IS+GANPI+ R+GVM+AV++V LK Sbjct: 100 LVQNVANKTNEEAGDGTTTATVLARPIAKEGFENISRGANPIEIRKGVMLAVESVKRQLK 159 Query: 362 AMSKPVTTPEEIAQVATISANGDRSIGDLISEAMKKVGKDGTITVKDGKTLHDELEVIEG 541 MSKPV T EEI QVATISANGD SIG LI+ AM +VGK+G ITVKDGKTL DELE+IEG Sbjct: 160 EMSKPVNTSEEIEQVATISANGDESIGKLIAAAMNRVGKNGVITVKDGKTLEDELEIIEG 219 Query: 542 MKFDRGYISPYFINTDKGAKCEFQDAFILFSEKKISSIQSLIPVLEMTHQQKKPLLIVAE 721 MKFDRGY+SPYFIN++KG K E+ DA +L+SEKKI ++P LE+ + QKKPL+I+AE Sbjct: 220 MKFDRGYVSPYFINSNKGPKVEYNDALVLYSEKKIYYASQVVPALELANSQKKPLVIIAE 279 Query: 722 DVDGEALTALVLNRLKLGLQVCAVKAPGFGDNRKNTLRDMATSTGGIVFGDEADTYKIEN 901 D DGE L+ LV+N+LK+GL V AVKAPGFG+ R N L DMA +TGG VF D+ + ++E+ Sbjct: 280 DYDGEPLSVLVVNKLKIGLPVVAVKAPGFGEYRTNALLDMAAATGG-VFEDDTNLVRLED 338 Query: 902 VQLHDLGRAQEILITKDDTLVMKGKGLQKDIQKRIDEIRDEIALSTSEY 1048 Q G+ E++ITKD TL++KGKG +I++RID+I++E+ +TS Y Sbjct: 339 CQAESFGQVGEVIITKDSTLLLKGKGDPNEIKQRIDQIKEELETATSNY 387
>sp|O74261|HSP60_CANAL Heat shock protein 60, mitochondrial precursor (60 kDa chaperonin) (Protein Cpn60) Length = 566 Score = 414 bits (1064), Expect = e-115 Identities = 210/350 (60%), Positives = 276/350 (78%), Gaps = 1/350 (0%) Frame = +2 Query: 2 RSAMLVGVDILADAVAVTMGPKGRNVILEKSWGSPKITKDGVTVAKDIELKDKFQNIGAK 181 R+A+L GV+ LADAV+VT+GPKGRNV++E+ +G+PKITKDGVTVAK I L+DKF+++GAK Sbjct: 29 RAALLKGVNTLADAVSVTLGPKGRNVLIEQQFGAPKITKDGVTVAKAITLEDKFEDLGAK 88 Query: 182 LVQDVANNTNEEAGDGTTAATVLARAIAKEGFEKISKGANPIDFRRGVMMAVDNVVASLK 361 L+Q+VA+ TNE AGDGTT+ATVL R+I E + ++ G NP+D RRG AV+ V+ L+ Sbjct: 89 LLQEVASKTNESAGDGTTSATVLGRSIFTESVKNVAAGCNPMDLRRGSQAAVEAVIEFLQ 148 Query: 362 AMSKPVTTPEEIAQVATISANGDRSIGDLISEAMKKVGKDGTITVKDGKTLHDELEVIEG 541 K +TT EEIAQVATISANGD+ IGDL++ AM+KVGK+G ITVK+GKTL DELEV EG Sbjct: 149 KNKKEITTSEEIAQVATISANGDKHIGDLLANAMEKVGKEGVITVKEGKTLEDELEVTEG 208 Query: 542 MKFDRGYISPYFINTDKGAKCEFQDAFILFSEKKISSIQSLIPVLEMTHQQKKPLLIVAE 721 MKFDRG+ISPYFI K K EF++ IL SEKKISSIQ ++P LE+++Q ++PLLI+AE Sbjct: 209 MKFDRGFISPYFITNTKTGKVEFENPLILLSEKKISSIQDILPSLELSNQTRRPLLIIAE 268 Query: 722 DVDGEALTALVLNRLKLGLQVCAVKAPGFGDNRKNTLRDMATSTGGIVFGDEADTYKIEN 901 DVDGEAL A +LN+L+ +QVCAVKAPGFGDNRKNTL D+A +GG VF +E D K EN Sbjct: 269 DVDGEALAACILNKLRGQVQVCAVKAPGFGDNRKNTLGDIAILSGGTVFTEELD-IKPEN 327 Query: 902 VQLHDLGRAQEILITKDDTLVMKGKGLQKDIQKRIDEIRDEIA-LSTSEY 1048 + LG A + ITK+DT+++ G+G + +++ R ++IR IA + T+EY Sbjct: 328 ATIEQLGSAGAVTITKEDTVLLNGEGSKDNLEARCEQIRSVIADVHTTEY 377
>sp|Q9VPS5|CH60B_DROME Probable 60 kDa heat shock protein homolog 1, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (CPN60) (Heat shock protein 60) (HSP-60) Length = 648 Score = 412 bits (1060), Expect = e-115 Identities = 204/343 (59%), Positives = 268/343 (78%) Frame = +2 Query: 2 RSAMLVGVDILADAVAVTMGPKGRNVILEKSWGSPKITKDGVTVAKDIELKDKFQNIGAK 181 R+ M+ GVDILADAVAVTMGPKGR+VI+E+ W SPKITKDG TVA+ I LKD+ N+GAK Sbjct: 31 RAMMIRGVDILADAVAVTMGPKGRSVIVERPWTSPKITKDGFTVARSIALKDQHMNLGAK 90 Query: 182 LVQDVANNTNEEAGDGTTAATVLARAIAKEGFEKISKGANPIDFRRGVMMAVDNVVASLK 361 LVQDVA+NTNE AGDGTT ATVLARAIAKEGF +I+ GANP++ RRGVM+AVD V LK Sbjct: 91 LVQDVADNTNESAGDGTTTATVLARAIAKEGFNQITMGANPVEIRRGVMLAVDVVKDKLK 150 Query: 362 AMSKPVTTPEEIAQVATISANGDRSIGDLISEAMKKVGKDGTITVKDGKTLHDELEVIEG 541 MSK V T EEI QVAT+SANGD IG LI EA KVG GTITVKDGK L DEL +I+G Sbjct: 151 EMSKAVETREEIQQVATLSANGDTEIGRLIGEATDKVGPRGTITVKDGKRLKDELNIIQG 210 Query: 542 MKFDRGYISPYFINTDKGAKCEFQDAFILFSEKKISSIQSLIPVLEMTHQQKKPLLIVAE 721 ++FD GY+SP+F+N+ KG+K EF +A ++ S KKI+ + ++ LE + +Q++PL+I+AE Sbjct: 211 LRFDNGYVSPFFVNSSKGSKVEFANALVMISLKKITGLSQIVKGLEQSLRQRRPLIIIAE 270 Query: 722 DVDGEALTALVLNRLKLGLQVCAVKAPGFGDNRKNTLRDMATSTGGIVFGDEADTYKIEN 901 D+ GEAL ALVLN+L+LGLQVCAVK+P +G +RK + D++ +TG +FGD+ + K+E Sbjct: 271 DISGEALNALVLNKLRLGLQVCAVKSPSYGHHRKELIGDISAATGATIFGDDINYSKMEE 330 Query: 902 VQLHDLGRAQEILITKDDTLVMKGKGLQKDIQKRIDEIRDEIA 1030 +L DLG+ E +I+KD T++++GK ++ RI +I+DE+A Sbjct: 331 AKLEDLGQVGEAVISKDSTMLLQGKPKTGLLEMRIQQIQDELA 373
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 110,407,696
Number of Sequences: 369166
Number of extensions: 2237087
Number of successful extensions: 8137
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7280
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7582
length of database: 68,354,980
effective HSP length: 111
effective length of database: 47,849,395
effective search space used: 11388156010
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail