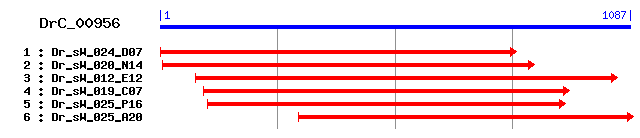
DrC_00956
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00956 (1087 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P53990|K0174_HUMAN Protein KIAA0174 (Putative MAPK-activ... 252 1e-66 sp|P53843|YN05_YEAST Hypothetical 34.5 kDa protein in PIK1-... 103 1e-21 sp|Q12446|LAS17_YEAST Proline-rich protein LAS17 41 0.007 sp|Q91YD9|WASL_MOUSE Neural Wiskott-Aldrich syndrome protei... 37 0.098 sp|O95466|FMNL_HUMAN Formin-like 1 protein (Formin-like pro... 36 0.22 sp|Q5R789|ZCHC8_PONPY Zinc finger CCHC domain containing pr... 35 0.37 sp|Q9JL26|FMNL_MOUSE Formin-like 1 protein (Formin-related ... 35 0.37 sp|Q8IN94|OSA_DROME Trithorax group protein osa (Eyelid pro... 35 0.49 sp|O43313|Y0431_HUMAN Hypothetical protein KIAA0431 34 0.64 sp|Q95107|WASL_BOVIN Neural Wiskott-Aldrich syndrome protei... 34 0.83
>sp|P53990|K0174_HUMAN Protein KIAA0174 (Putative MAPK-activating protein PM28) Length = 364 Score = 252 bits (643), Expect = 1e-66 Identities = 154/372 (41%), Positives = 211/372 (56%), Gaps = 42/372 (11%) Frame = +2 Query: 23 MFSSKPNYNKLKTNLRIVVNRLKLLEKKNTEIALKSRKEIADYLTSAKYDRARIKVEMII 202 M S +L+ NLR+V+NRLKLLEKK TE+A K+RKEIADYL + K +RARI+VE II Sbjct: 1 MLGSGFKAERLRVNLRLVINRLKLLEKKKTELAQKARKEIADYLAAGKDERARIRVEHII 60 Query: 203 RQDYLVEVYELIEMFCDLLLARFGLIECQKEIDPGLEEQIATLIWVTPRLQTDVQELKLI 382 R+DYLVE E++E++CDLLLARFGLI+ KE+D GL E ++TLIW PRLQ++V ELK++ Sbjct: 61 REDYLVEAMEILELYCDLLLARFGLIQSMKELDSGLAESVSTLIWAAPRLQSEVAELKIV 120 Query: 383 TDQLHLKYGREYGLACQSNSLNNVNEKVMEKLSIKAPPKRLVEQYLVEIAKSFDVPYEPE 562 DQL KY +EYG C++N + VN+++M KLS++APPK LVE+YL+EIAK+++VPYEP+ Sbjct: 121 ADQLCAKYSKEYGKLCRTNQIGTVNDRLMHKLSVEAPPKILVERYLIEIAKNYNVPYEPD 180 Query: 563 EDIDPPASNWNPGEFYAPLPPNLIDF-MGEKPPLINPNAYXXXXXXXXXXXXXXXXXXXX 739 + A PG + +LID + P Sbjct: 181 SVVMAEAP---PG-----VETDLIDVGFTDDVKKGGPGRGGSGGFTAPVGGPDGTVPMPM 232 Query: 740 XXXXXISNAPYPYP-QKKPLNDNEAPM---------------SLPPKYEEFD-------- 847 +N P+ YP K P + N PM + PP YE D Sbjct: 233 PMPMPSANTPFSYPLPKGPSDFNGLPMGTYQAFPNIHPPQIPATPPSYESVDDINADKNI 292 Query: 848 --------GSSSQESA--PPLPPNGPNDLFLPEIPSDQNNTNPPPGSGS-------VDFD 976 G + SA P P + ++ LPE+PS +T P +G+ +DFD Sbjct: 293 SSAQIVGPGPKPEASAKLPSRPADNYDNFVLPELPS-VPDTLPTASAGASTSASEDIDFD 351 Query: 977 DLNRRFEELKRR 1012 DL+RRFEELK++ Sbjct: 352 DLSRRFEELKKK 363
>sp|P53843|YN05_YEAST Hypothetical 34.5 kDa protein in PIK1-POL2 intergenic region Length = 298 Score = 103 bits (256), Expect = 1e-21 Identities = 70/216 (32%), Positives = 120/216 (55%), Gaps = 8/216 (3%) Frame = +2 Query: 50 KLKTNLRIVVNRLKLLEKKNTEIALKSRKEIADYLTSAKYDRARIKVEMIIRQDYLVEVY 229 KLKT L++ + RL+ ++K IA +SR+++A L + K +A +VE +I D +E+ Sbjct: 11 KLKTCLKMCIQRLRYAQEKQQAIAKQSRRQVAQLLLTNKEQKAHYRVETLIHDDIHIELL 70 Query: 230 ELIEMFCDLLLARFGLIE--------CQKEIDPGLEEQIATLIWVTPRLQTDVQELKLIT 385 E++E++C+LLLAR +I ++ +D G+ E I +LI+ +V+EL + Sbjct: 71 EILELYCELLLARVQVINDISTEEQLVKEHMDDGINEAIRSLIYAI-LFVDEVKELSQLK 129 Query: 386 DQLHLKYGREYGLACQSNSLNNVNEKVMEKLSIKAPPKRLVEQYLVEIAKSFDVPYEPEE 565 D + K E+ ++ + +V EK+++K S P + LV+ YL EIAK++DVPY E Sbjct: 130 DLMAWKINVEFVNGVIADHI-DVPEKIIKKCSPSVPKEELVDLYLKEIAKTYDVPYSKLE 188 Query: 566 DIDPPASNWNPGEFYAPLPPNLIDFMGEKPPLINPN 673 + +S+ +F P ++ D EKP L N Sbjct: 189 NSLSSSSSNISSDFSDP-SGDIEDNDEEKPILALDN 223
>sp|Q12446|LAS17_YEAST Proline-rich protein LAS17 Length = 633 Score = 40.8 bits (94), Expect = 0.007 Identities = 23/61 (37%), Positives = 32/61 (52%) Frame = +2 Query: 770 YPYPQKKPLNDNEAPMSLPPKYEEFDGSSSQESAPPLPPNGPNDLFLPEIPSDQNNTNPP 949 +P PL + AP+ PP++ S Q +AP P + P +PEIPS Q+ TNP Sbjct: 234 HPKHSLPPLPNQFAPLPDPPQHN----SPPQNNAPSQPQSNPFPFPIPEIPSTQSATNPF 289 Query: 950 P 952 P Sbjct: 290 P 290
>sp|Q91YD9|WASL_MOUSE Neural Wiskott-Aldrich syndrome protein (N-WASP) Length = 501 Score = 37.0 bits (84), Expect = 0.098 Identities = 23/78 (29%), Positives = 30/78 (38%) Frame = +2 Query: 776 YPQKKPLNDNEAPMSLPPKYEEFDGSSSQESAPPLPPNGPNDLFLPEIPSDQNNTNPPPG 955 YP P + AP PP S+ PP PP P P +PSD ++ P P Sbjct: 340 YPPPPPALPSSAPSGPPPPPPPSMAGSTAPPPPPPPPPPPGPPPPPGLPSDGDHQVPAPS 399 Query: 956 SGSVDFDDLNRRFEELKR 1009 D R +LK+ Sbjct: 400 GNKAALLDQIREGAQLKK 417
>sp|O95466|FMNL_HUMAN Formin-like 1 protein (Formin-like protein) (Leukocyte formin) (CLL-associated antigen KW-13) Length = 1100 Score = 35.8 bits (81), Expect = 0.22 Identities = 24/64 (37%), Positives = 28/64 (43%), Gaps = 1/64 (1%) Frame = +2 Query: 764 APYPYPQKKPL-NDNEAPMSLPPKYEEFDGSSSQESAPPLPPNGPNDLFLPEIPSDQNNT 940 AP P P L + EAP S PP+ GS APPLP + P P P + Sbjct: 541 APPPPPPLPGLPSPQEAPPSAPPQAPPLPGSPEPPPAPPLPGDLPPPPPPPPPPPGTDGP 600 Query: 941 NPPP 952 PPP Sbjct: 601 VPPP 604
>sp|Q5R789|ZCHC8_PONPY Zinc finger CCHC domain containing protein 8 Length = 704 Score = 35.0 bits (79), Expect = 0.37 Identities = 33/99 (33%), Positives = 38/99 (38%), Gaps = 19/99 (19%) Frame = +2 Query: 773 PYPQKKPLNDNEAP--MSLPPKYEEFDGSSSQES----------APPLPPNGPNDLFLPE 916 P QKK N +P M L E GS S ES PPLP P +F P Sbjct: 426 PKKQKKESNSAGSPADMELDSDMEVPHGSQSSESFQFQPPLPPDTPPLPRGTPPPIFTPP 485 Query: 917 IPSDQNNTNPP------PGSGSVDFDDLN-RRFEELKRR 1012 +P P SG+VD D L EE +RR Sbjct: 486 LPKGTPPLTPSDSPQTRTASGAVDEDALTLEELEEQQRR 524
>sp|Q9JL26|FMNL_MOUSE Formin-like 1 protein (Formin-related protein) Length = 1094 Score = 35.0 bits (79), Expect = 0.37 Identities = 24/69 (34%), Positives = 29/69 (42%), Gaps = 4/69 (5%) Frame = +2 Query: 758 SNAPYPYPQKKPL----NDNEAPMSLPPKYEEFDGSSSQESAPPLPPNGPNDLFLPEIPS 925 S P P P PL + EAP S PP G + APPLP + P P + + Sbjct: 536 SPPPPPPPPPPPLPNLQSQQEAPPSAPPLAPPLPGCAEPPPAPPLPGDLPPPPPPPPLGT 595 Query: 926 DQNNTNPPP 952 D PPP Sbjct: 596 DGPVPPPPP 604
>sp|Q8IN94|OSA_DROME Trithorax group protein osa (Eyelid protein) Length = 2716 Score = 34.7 bits (78), Expect = 0.49 Identities = 23/71 (32%), Positives = 30/71 (42%), Gaps = 8/71 (11%) Frame = +2 Query: 773 PYPQKKPLNDNEAPMSLPPKYEEFDGSSSQE-SAPPLPPNGPNDLFLPEIPSDQNN---- 937 P Q++ AP + PP G+++ S PP P N N+ P I Q N Sbjct: 8 PQTQQQQQGGAPAPAATPPSAGAAPGAATPPTSGPPTPNNNSNNGSDPSIQQQQQNVAPH 67 Query: 938 ---TNPPPGSG 961 PPPGSG Sbjct: 68 PYGAPPPPGSG 78
Score = 30.8 bits (68), Expect = 7.0
Identities = 23/71 (32%), Positives = 28/71 (39%), Gaps = 9/71 (12%)
Frame = +2
Query: 767 PYPYPQ-----KKPLNDNEAPMSLPPKYEEFDGSSS----QESAPPLPPNGPNDLFLPEI 919
PY Y Q K P P PP+ GS + Q P PP GP P +
Sbjct: 160 PYRYGQPLPGGKPPQQQQPHPQQQPPQQPGPGGSPNRPPQQRYIPGQPPQGPT----PTL 215
Query: 920 PSDQNNTNPPP 952
S ++NPPP
Sbjct: 216 NSLLQSSNPPP 226
>sp|O43313|Y0431_HUMAN Hypothetical protein KIAA0431 Length = 667 Score = 34.3 bits (77), Expect = 0.64 Identities = 17/54 (31%), Positives = 28/54 (51%) Frame = +2 Query: 755 ISNAPYPYPQKKPLNDNEAPMSLPPKYEEFDGSSSQESAPPLPPNGPNDLFLPE 916 ++N P P P + L +E + L P +E+ GS++ + PP P L LP+ Sbjct: 91 LNNQPIPRPDTQELEASE--IKLEPSFEDSCGSNTDKQTLTTPPRYPQKLLLPK 142
>sp|Q95107|WASL_BOVIN Neural Wiskott-Aldrich syndrome protein (N-WASP) Length = 505 Score = 33.9 bits (76), Expect = 0.83 Identities = 24/79 (30%), Positives = 30/79 (37%), Gaps = 1/79 (1%) Frame = +2 Query: 776 YPQKKPLNDNEAPMSLPPKYEEFDGSSS-QESAPPLPPNGPNDLFLPEIPSDQNNTNPPP 952 YP P + AP PP S S PP PP P P +PSD ++ P P Sbjct: 343 YPPPLPALPSSAPSGPPPPPPPLSVSGSVAPPPPPPPPPPPGPPPPPGLPSDGDHQVPTP 402 Query: 953 GSGSVDFDDLNRRFEELKR 1009 D R +LK+ Sbjct: 403 AGSKAALLDQIREGAQLKK 421
Score = 31.6 bits (70), Expect = 4.1
Identities = 23/63 (36%), Positives = 26/63 (41%)
Frame = +2
Query: 764 APYPYPQKKPLNDNEAPMSLPPKYEEFDGSSSQESAPPLPPNGPNDLFLPEIPSDQNNTN 943
AP P P + P AP PP S APP PPN LP +PS +
Sbjct: 309 APPPPPSRAP---TAAPPPPPP-------SRPGVGAPPPPPNRMYPPPLPALPSSAPSGP 358
Query: 944 PPP 952
PPP
Sbjct: 359 PPP 361
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 119,154,147
Number of Sequences: 369166
Number of extensions: 2567067
Number of successful extensions: 8427
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7239
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8257
length of database: 68,354,980
effective HSP length: 112
effective length of database: 47,664,660
effective search space used: 11868500340
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail